Temporary protein-protein bonds are essential for processes including enzymatic reactions, antibody binding, and response to medication. Being able to accurately characterize these bonds is important for testing the performance of potential therapies, but currently available methods for doing this have limited capacity to either provide information at the single bond level, or to test large numbers of bonds.
Researchers at the Centre National de la Recherche Scientifique (CNRS) and their colleagues have now presented a more accessible method to measure the strength and duration of protein-protein bonds under similar loads to those they would experience inside our bodies. The method uses sound waves to pull bonded proteins apart and DNA leashes to keep the two proteins close together so that they can re-bond after their connection is ruptured. This innovation allows the same protein bonds to be re-tested up to 100 times, providing valuable insights about how bond strength changes as molecules age. This ability could provide new information on the half-life of drugs or antibodies.
Reporting on their work in Biophysical Journal (“Combining DNA scaffolds and acoustic force spectroscopy to characterize individual protein bonds”), senior author Laurent Limozin, PhD, a CNRS biophysicist, and colleagues, stated, “This proof-of-principle is established for two protein bonds of biomedical interest, opening promising perspectives for biotechnological and medical studies.”
The binding properties of biomolecules, which govern biological phenomena, are relevant for evaluating therapeutics, the team explained, but new experimental tools are needed to characterize, in a multiplexed manner, protein bond rupture under force. “While bulk measurements performed on populations of molecules remain the standard characterization techniques, single-molecule force spectroscopy (SMFS) on individual pairs of interacting partners has emerged as a powerful complementary strategy because it uniquely gives access to individual bond response to force.”
For their newly reported study, Limozin and colleagues combined, for the first time, they claimed, DNA scaffolds with acoustic force spectroscopy (AFS) to measure the force response of individual biomolecular complexes. Acoustic force spectroscopy allows many molecular pairs to be tested simultaneously, and the DNA scaffolds allowed repeated testing of the same bonds.
“We wanted to propose a method that is sufficiently modular to apply to different type of bonds, that has a reasonable throughput, and that reaches high molecular precision that is currently only available with very refined techniques, like optical or magnetic tweezers, that are often difficult to grasp for non-specialists,” Limozin said.
During acoustic force spectroscopy, pairs of bonded proteins are tested inside a liquid-filled chamber. The proteins are restrained by DNA scaffolding such that one strand of DNA attaches the first protein to the bottom of the chamber, while another strand attaches the second protein to a small silica bead. When the researchers blast the chamber with a soundwave, the wave’s force pulls the silicon bead—and the protein it’s attached to—away from the bottom of the chamber. If the force is strong enough, this pulling action ruptures the bond between the two proteins.
For the new method, a third strand of DNA acts as a leash to keep the proteins close together after their bond is ruptured. “… we introduce the combination of a modular DNA scaffold, namely junctured-DNA (J-DNA), with AFS, an emerging parallel method that potentially offers a large dynamic range of fast force application,” the scientists wrote. “The probed protein complex is attached to both the surface of the flow cell and a bead via kilobase pairlong DNA shanks connected by the leash.”
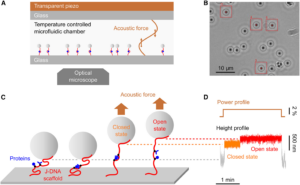
“The originality of our method is that in addition to these two strands on each side, in the middle you have this leash that connects the two strands and keeps the proteins together upon rupture,” explained Limozin. “Without this leash, the detachment would be irreversible, but this allows you to repeat the measurement almost as many times as you wish.”
As a proof of concept, the research team used the technique to characterize two single-molecule interactions of biomedical interest— the bond between proteins and rapamycin, an immunosuppressive drug, and the bond between a single-domain antibody and an HIV-1 antigen. “In addition to its interest for benchmarking, rapamycin has an important biomedical relevance, being one of the earliest modulators of protein-protein interaction to have reached the clinic,” the researchers commented. The researchers observed the cycles of bonding and rupturing using a microscope. They also compared their results with those of well-established techniques, such as magnetic tweezers, to ensure their accuracy.
Being able to test the same protein-protein bond multiple times is important for exploring variation between molecularly identical pairs. It also allows researchers to examine how these interactions change as the molecules age, which could be important for determining the half-life of drugs or antibodies. “ … combining AFS and J-DNA has the potential to both answer fundamental questions and open the way to systematic chemomechanical characterization of biomedically relevant interactions,” they noted.
“With this tool we have a way to go deeper and really probe experimentally ideas about molecular heterogeneity and molecular aging,” said Limozin. “We, and others, suspect that characterizing these properties will be very useful for designing future therapeutics that will need to work in situations where mechanical forces are involved.”



