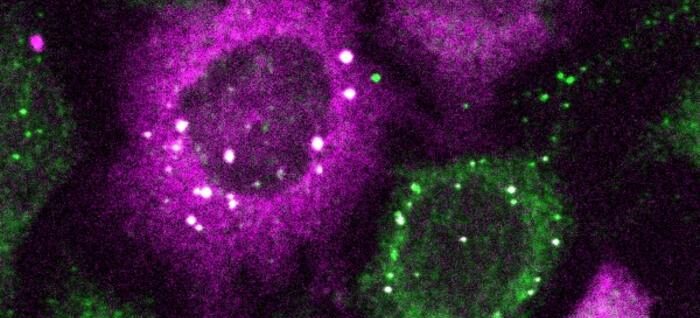
CluMPS—Clusters Magnified by Phase Separation—is a fluorescent reporter strategy for detecting protein clusters. It promises to facilitate the visualization of protein clusters that are too small to detect with conventional fluorescence microscopy. Such protein clusters have been implicated in diseases such as Alzheimer’s and are, consequently, of abiding concern to researchers.
The “phase separation” in CluMPS refers to the generation of large, quantifiable biomolecular condensates, which are sometimes called membrane-less organelles. They are, in essence, blobs of phase-separated liquids that serve to concentrate collections of proteins and nucleic acids. They separate out of the cytoplasm rather like oil droplets separate out of water.
CluMPS was developed by a research team based at the University of Pennsylvania School of Engineering and Applied Science. The team, which was led by Lukasz J. Bugaj, PhD, assistant professor of engineering, presented its findings in Cell Systems, in a paper titled, “Simple visualization of submicroscopic protein clusters with a phase-separation-based fluorescent reporter.”
The paper’s authors noted that CluMPS incorporates a fluorescent reporter that can detect and visually amplify even small clusters of a binding partner. Moreover, the authors asserted that CluMPS provides a “powerful yet straightforward approach” to observe higher-order protein assembly in its native cellular context.
“We use computational modeling and optogenetic clustering to demonstrate that CluMPS can detect small oligomers and behaves rationally according to key system parameters,” the authors detailed. “CluMPS detected small aggregates of pathological proteins where the corresponding GFP fusions appeared diffuse. CluMPS also detected and tracked clusters of unmodified and tagged endogenous proteins, and orthogonal CluMPS probes could be multiplexed in cells.”
Normally, detecting protein clusters requires laborious techniques. “But with CluMPS,” Bugaj said, “you don’t need anything beyond the standard lab microscope.”
The tool fuses with the target protein to form condensates orders of magnitude larger than the protein clusters themselves that resemble the colorful blobs in a lava lamp. “We think the simplicity of the approach is one of its main benefits,” Bugaj remarked. “You don’t need specialized skills or equipment to quickly see whether there are small clusters in your cells.”
For treating diseases like Alzheimer’s, ALS, and even cancer, gaining the ability to detect such tiny protein clusters promises to be a foundational advancement, allowing researchers to determine whether drugs actually eliminate disease-causing clusters of a target protein in a cell.
“You need a very clear signal” to know whether a treatment worked, Bugaj said. “It’s very obvious when you have a gigantic cluster, but if you have small clusters, it is much harder. Now we can amplify that signal and see which drugs actually dissolve the clusters.”
In addition to providing new avenues for drug discovery, CluMPS will permit researchers to understand the functioning of proteins in new ways, leading to a deeper, more sophisticated rendering of cells themselves. “There’s an entire landscape of protein clustering that’s happening at the small scale,” Bugaj pointed out. “That’s important, but we just don’t know about it yet.”
One of the challenges that CluMPS overcomes is that lightwaves themselves are larger than the smallest protein clusters, making it very hard to see such clusters without specialized techniques. “The wavelength of blue light is about 400 nanometers,” Bugaj said. “You can’t actually resolve the location of anything smaller than half that wavelength with a conventional microscope,” rendering protein clusters tens of nanometers wide all but invisible.
To develop CluMPS, Bugaj and his laboratory partnered with Elizabeth Rhoades, PhD, professor of chemistry at Penn Arts and Sciences, whose laboratory helped validate that CluMPS did indeed detect target protein clusters instead of generating false positives.
Thomas R. Mumford, a doctoral student in the Bugaj laboratory and the paper’s lead author, noted, “It was crucial to characterize how underlying features of protein clusters interacted with CluMPS to trigger condensation” and thereby enable future users of the technology to understand precisely how it works.