The SARS-CoV-2 virus is continuously evolving. The ability to detect the evolving variants specifically and efficiently is a key step in combating the progress of the COVID-19 pandemic that contrary to popular notions, is far from over.
Scientists at Sichuan University in Chengdu and Tsinghua University in Beijing have developed an inexpensive enzymatic colorimetric assay that can simultaneously detect the presence of SARS-CoV-2 and discriminate among mutations specific to alpha, beta, and gamma variants. Each test costs only about $0.30 and provides answers within 30 minutes.
The steps of this assay are detailed in an article in the journal Nature Biomedical Engineering titled, “A paper-based assay for the colorimetric detection of SARS-CoV-2 variants at single-nucleotide resolution.”
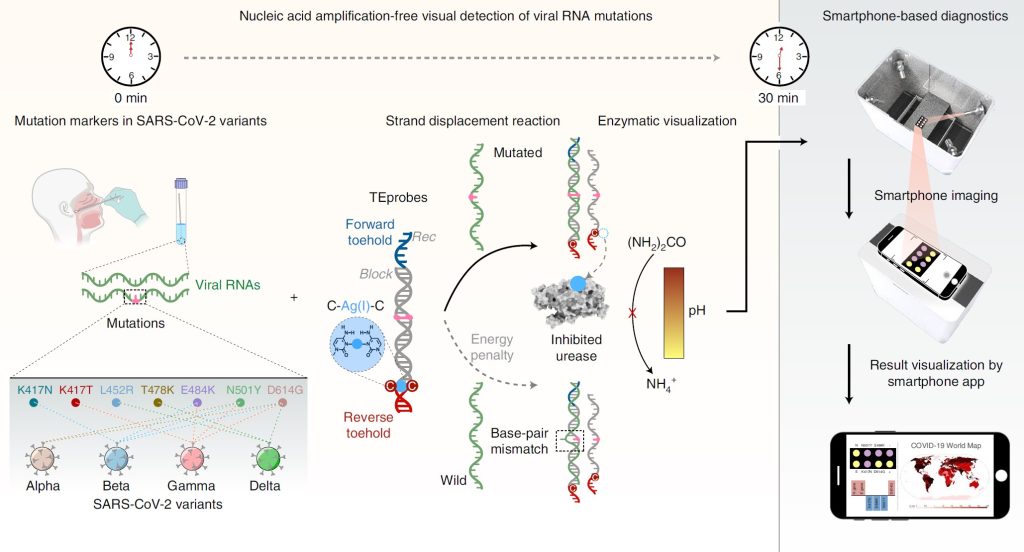
“The assay, which we integrated into foldable paper strips, leverages nucleic acid strand-displacement reactions, the thermodynamic energy penalty associated with single-base-pair mismatches, and the metal-ion-controlled enzymatic cleavage of urea to amplify the recognition of viral RNAs for the colorimetric readout of changes in pH via a smartphone,” the authors noted.
The lack of affordable and scalable diagnostic tools to detect SARS-CoV-2 variants is impeding population-wide detection of infections and epidemiological surveillance. As new variants arise, doubts around viral transmission, pathogenicity, and vaccine effectiveness can only be addressed through prompt and accurate detection of variants, globally.
Ideally, screens for SARS-CoV-2 variants should be able to detect single nucleotide mutations in viral RNAs, provide quick answers, and adopt a flexible multiplexed approach that enables the identification of multiple existing variants as well as emerging variants with simple modifications.
“The need for scalable SARS-CoV-2 tests that allow variants to be resolved has motivated our efforts to explore single nucleotide-resolved viral RNA detection strategies,” the authors noted.
The authors have named the new assay MARVE for “multiplexed, nucleic-acid-amplification-free, single-nucleotide-resolved viral evolution.” The test does not require the amplification of viral nucleic acids using PCR or RT-PCR and yet detects viral RNAs at single nucleotide resolution. To accomplish this, the researchers used a programmable enzyme-induced nucleic acid strand displacement process to detect individual viral RNAs in the sample. This eliminates the need for nucleic acid amplification and reduces the cost of production of the test and the turnaround time for conclusive readouts.
The strand displacement reaction is made possible through a double-stranded DNA probe, which the authors call the toehold exchange DNA probe (TEprobe), designed to recognize mutations in SARS-CoV-2 variants (alpha, beta, gamma, and delta). The TEprobe consists of a pair of terminal single-stranded overhangs that form the forward and reverse toeholds. Designed sequences of the forward and reverse toeholds enable the TEprobe to precisely control the net thermodynamic energy of the strand displacement reaction induced by viral RNAs in the sample. The authors successfully distinguished mutant and wild viral RNAs through the strand displacement reaction based on the thermodynamic energy penalty from a single mismatch in the sequence.
Recognition of viral RNAs releases a silver ion that inhibits urease—an enzyme that breaks urea down into ammonia, carbon dioxide, and water. The release of the metal ion thereby controls the cleavage of urea, the level of resulting ammonium ions, and the pH. Known mutations in SARS-CoV-2 variants can therefore be identified visually using pH indicators.
“To demonstrate the simplicity, portability, and multiplexing capability of MARVE, we measured SARS-CoV-2 and its five key variants in an integrated testing paper using a smartphone. We also developed a smartphone application to guide the diagnosis, and to visualize and record the test results for clinical and cold-chain food samples, facilitating on-site profiling of SARS-CoV-2 variants by minimally trained personnel,” the authors noted.
The team tested the assay on 50 throat swab samples and simultaneously detected the presence of SARS-CoV-2 and mutations specific to the alpha, beta, and gamma variants of SARS-CoV-2 variants. They showed that their readouts match results obtained through real-time quantitative polymerase chain reaction and RNA sequencing.
This customizable and inexpensive paper-based assay that detects the COVID-19 virus and its variants can aid viral surveillance and population-wide diagnosis and screening without the need for complex and time-consuming laboratory tests.



