Proper cellular function needs accurate mechanisms of transporting newly synthesized proteins to their respective cellular niches. In a collaborative study, scientists have discovered how newborn proteins are transported to the endoplasmic reticulum (ER)—a cytoplasmic network of membranous tunnels that leads into the nuclear membrane.
“A longstanding open question that is fundamental to life is how cells manage to produce proteins and transport them during their synthesis to their correct cellular destinations,” said Elke Deuerling, PhD, a professor of molecular microbiology at the University of Konstanz, Germany, and a senior author of this study.
In an article titled, “Mechanism of signal sequence handover from NAC to SRP on ribosomes during ER-protein targeting,” published in the journal Science, Deuerling and her team parse out the molecular mechanism of how a protein complex called NAC (nascent-polypeptide-associated complex) that reversibly binds to eukaryotic ribosomes, interacts with newly synthesized polypeptide chains as they emerge from the ribosomal tunnel, and hands over hydrophobic ER targeting signal sequences encoded in the polypeptide, to the signal recognition particle (SRP), a protein-RNA transporter complex that targets proteins to the ER.
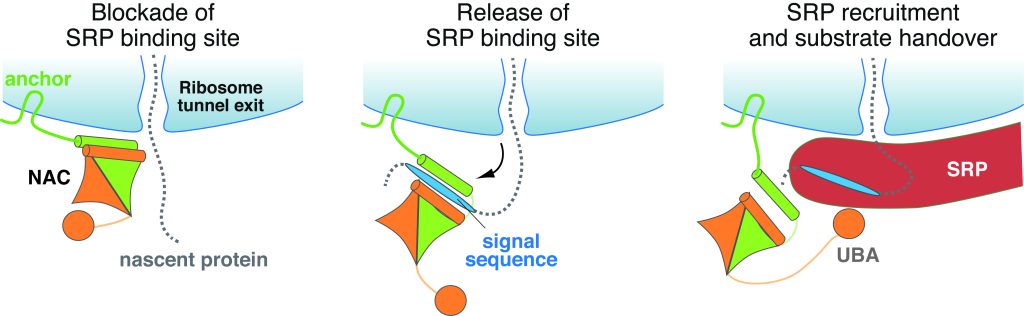
NAC ensures that only proteins destined for the ER are passed on to the SRP. The authors showed that nascent proteins with destinations other than the ER are denied access to the SRP by NAC, which acts as a gatekeeper in the process.
“We knew already that two factors (called NAC and SRP) together with a signal sequence in the nascent protein being synthesized on the ribosome are of decisive importance for the transport of a protein to the endoplasmic reticulum, but we did not understand how they interplay with each other to regulate and ensure correct protein targeting,” said Deuerling. “A powerful international cooperation among experts in this field solved this question by unraveling the molecular details of the NAC-SRP-nascent protein interplay on ribosomes.”
Earlier studies had established NAC interacts with newly synthesized proteins at the ribosomal exit and competes with the SRP to stop proteins that are meant for the cytosol or mitochondria from ending up in the ER. Yet, SRP can also bind nonspecifically to ribosomes that have no ER signal and carry it to the ER. It was unclear until now, how NAC prevents SRP from binding to just any ribosome and how this antagonistic interaction is overcome by ER targeting signals so that only the correct ribosomes are transported to the ER.
“Uncontrolled, SRP would bind to any ribosome close by and then transport it to the ER, regardless of whether or not a protein with that destination is currently being produced. This would result in countless misdeliveries that would severely impair the function and viability of the cell,” said Deuerling.
Deuerling’s team collaborated with scientists from ETH Zurich in Switzerland, MRC Laboratory of Molecular Biology (MRC-LMB) in Cambridge, U.K., and the California Institute of Technology, to solve this puzzle. They simulated the cellular process in vitro by mixing purified ribosomes, NAC and SRP, in a test tube. They then quickly froze the mixture to -150°C and examined the sample by cryoelectron microscopy.
This enabled structural biologists and co-authors of the study, Ahmad Jomaa, PhD, and Viswanathan Chandrasekaran, PhD, to determine how NAC binds to ribosomes before and after the ER signal-carrying newborn peptide is transferred to the SRP. This clarified NAC’s gatekeeper mechanism, but the transition between the states remained unclear.
“The transition is a highly dynamic process that cannot be visualized by cryoelectron microscopy,” said Martin Gamerdinger, PhD, a lead author from the University of Konstanz.
High-resolution biochemical binding studies conducted by Gamerdinger and his team—doctoral researchers Annalena Wallisch and Zeynel Ulusoy—showed the type of protein being synthesized controls the interaction of NAC with ribosomes. Hao-Hsuan Hsieh, PhD, conducted experiments on binding strength between the components, which together with computer-assisted 3D reconstruction, allowed the team to decipher the details of NAC’s sorting function.
The mechanistic model that results from the study proposes NAC uses two domains with opposing effects to control SRP access. NAC’s core globular domain prevents SRP from binding to ribosomes that do not bear the ER signal, whereas a flexibly attached domain temporarily grabs SRP to permit scanning of newborn peptides. When a peptide bearing an ER-targeting signal emerges from the ribosomal tunnel, it destabilizes NAC’s globular domain allowing SRP to bind to the correct chain and carry it off to the ER.
“Our study reveals the molecular function of NAC as a gatekeeper, granting SRP only access for those nascent proteins whose destination is the endoplasmic reticulum,” said Deuerling.
Errors in protein sorting are the mechanistic basis for many diseases. Deuerling believes this study that clarifies a vital cellular process will help advance studies in several fields of cell biology and life sciences.

Deuerling and her collaborators, Nenad Ban, PhD, from ETH Zurich, Shu-ou Shan, PhD, from Caltech, and Ramanujan Hegde, PhD, from MRC-LMB, believe determining whether NAC play other regulatory roles at the ribosomal exit will need further experimentations.
“There is a whole ‘welcoming committee’ including NAC and SRP but also many other factors that bind to ribosomes during the synthesis of new proteins,” said Deuerling. “Understanding how these factors acting at the front-end of the protein production line are coordinated in a spatial and temporal manner to ensure proteome integrity and cell viability will be the next steps.”