It is common knowledge that Down syndrome—a neurodevelopmental and neurodegenerative disorder with hallmark physical features and mild to moderate cognitive impairment—occurs when individuals have an extra copy of chromosome 21 (trisomy 21). One in 700 babies in the United States is born with Down syndrome. Despite being the most common chromosomal disorder, it is not clear how the presence of the extra chromosome triggers abnormalities in cells.
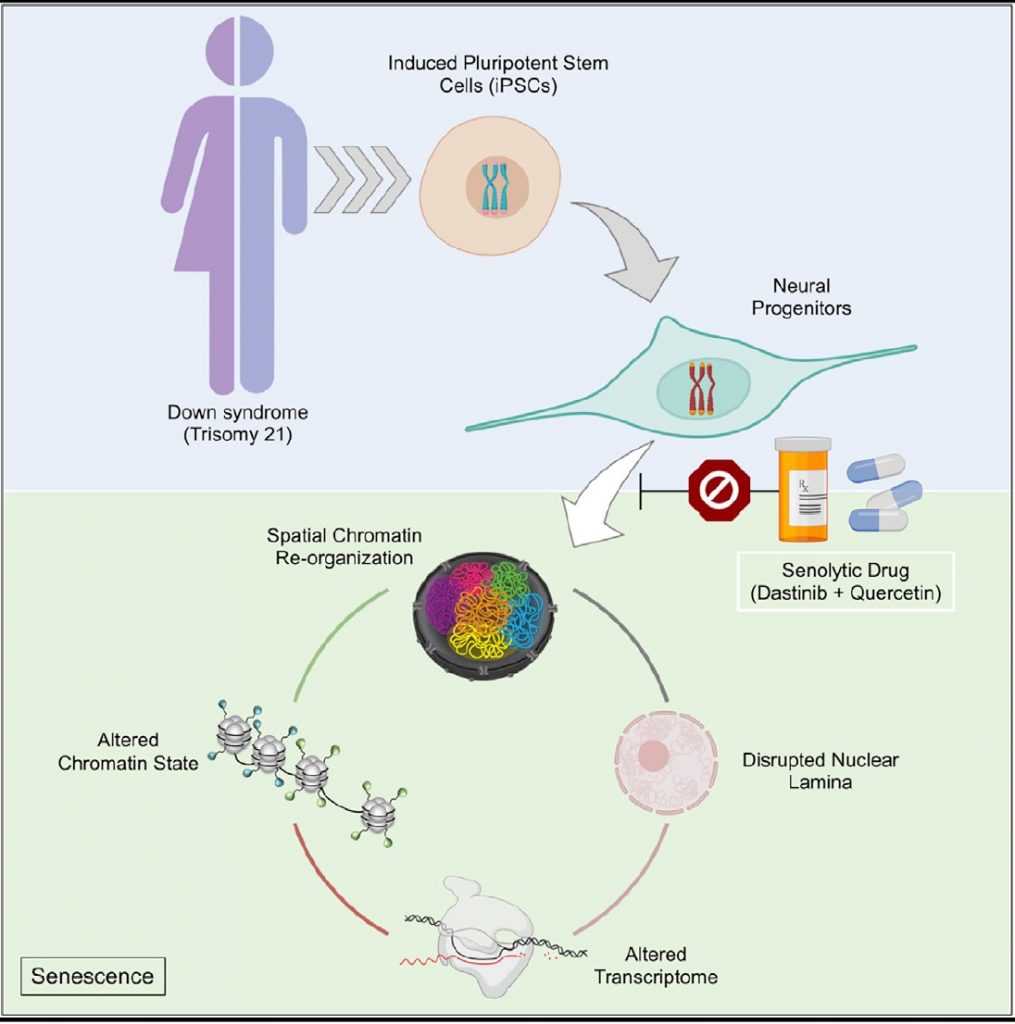
In a new study, scientists show that trisomy 21, like aging, disrupts the spatial organization of the genome, epigenome, and transcriptome in neural progenitor cells (NPCs)—precursor cells destined to specialize into neuronal cells. The authors also show, senolytic drugs—drugs that selectively kill off old cells to improve overall health—may reverse this nuclear disarray.
“The identification of the underlying mechanisms associated with neural progenitors opens new therapeutic avenues for Down syndrome. These approaches have not been explored in the context of Down syndrome, to our knowledge,” said Hiruy Meharena, PhD, assistant professor at the University of California, San Diego, who led the work as a postdoctoral fellow at the Alana Down Syndrome Center at the Massachusetts Institute of Technology (MIT).
These findings published in an article in the journal Cell Stem Cell on January 6, titled, “Down syndrome-induced senescence disrupts the nuclear architecture of neural progenitors.”
“First, through comprehensive analysis of the 3D genome in human trisomy 21 iPSCs and NPCs, they reveal specific chromatin changes in differentiated NPCs, but not in iPSCs, providing foundational data suggesting the effects of trisomy 21 are more impactful as cells differentiate,” said Bhattacharya. “Second, synthesis of the epigenome and transcriptome in these cells indicates trisomy 21 NPCs exhibit characteristics of senescent cells and that these characteristics can be reversed with senolytic drugs.”
Earlier transcriptomic analysis of tissues from individuals with Down syndrome had indicated trisomy 21 disrupts genome-wide transcription. “We were interested in identifying the mechanisms underlying the genome-wide transcriptional changes associated with Down syndrome and if the 3D-genome organization is involved in this process,” said Meharena.
To explore the effects of trisomy 21 on the molecular and cellular changes seen in Down syndrome, the researchers established iPSCs (induced pluripotent stem cells) and iPSC-derived forebrain NPCs from human cells of several individuals.
“We discovered that triplication of chromosome 21 induces global 3D-genome re-organization in neural progenitors and find that every chromosome becomes introverted—where each chromosome interacts more with itself than its neighboring chromosomes. This is similar to what has been observed in senescent cells, and thus we utilized senolytic drugs to ameliorate the genomic, molecular, and cellular dysfunctions observed in neural progenitors derived from individuals with Down syndrome,” said Meharena.
“This study illustrates the importance of asking fundamental questions about the underlying mechanisms of neurological disorders,” said senior author Li-Huei Tsai, the Picower professor of neuroscience, director of the Alana Center and of the Picower Institute for Learning and Memory at MIT.
Tsai said, “We didn’t begin this work expecting to see senescence as a translationally relevant feature of Down syndrome, but the data emerged from asking how the presence of an extra chromosome affects the architecture of all of a cell’s chromosomes during development.”

“We used Hi-C to interrogate the 3D-genome architecture and senolytic drugs to alleviate the phenotypes associated with trisomy 21,” said Meharena.
In addition to using Hi-C to explore the interactions within and between chromosomes, the team examined 3D chromosome architecture, metrics of DNA structure, gene accessibility, and expression in iPSCs and NPCs. They also examined differences in gene expression and their functional consequences in these developmental cells, such as changes in cell division and migration in 3D brain tissue cultures. The authors observed, although stem cells were not particularly different, NPCs were affected by trisomy 21 showing alterations in genomic organization, interaction, and expression, as well as deficits in proliferation and movement.
The similarities in genome structure, transcriptomic expression, and functional profiles between Down syndrome cells and senescent cells, spurred the team to test a combination of senolytic drugs—dasatinib and quercetin—to undo the abnormalities. The new approach improved gene accessibility and transcription, and also corrected deficits in cell migration and division.
However, these findings do not justify the use of these senolytic drugs in intervening with brain development in individuals with Down syndrome. Both drugs have significant side effects. For instance, dasatinib is used solely in cancer treatment when other treatment options have been exhausted. Safe senolytic drugs, once developed, could lead to new therapeutic options for Down syndrome.
The authors pointed out several limitations of their study including the fact that the iPSCs and NPCs used in the study were derived from a small number of individuals who died very young indicating a highly severe form of trisomy 21. Cells derived from older individuals with less severe expression of the disorder would be needed to estimate the range of the genomic disruption. In addition, although patient-derived iPSCs and NPCs provide a good model to study cellular function, they may not strictly adhere to the expression of the disease in patients.
“We would like to explore this phenomenon in vivo utilizing the available mouse models of Down syndrome,” said Meharena.
Bhattacharya added, “Future in-depth experiments to test the consequences of these drugs on specific neurodevelopmental cellular processes will be of great interest. Despite the limitations acknowledged by the authors, the analysis of human trisomy 21 PSCs enables the elucidation of the molecular impacts of trisomy 21 on different cell types. It will be interesting to see if progenitors and differentiated cells of other cell lineages that are affected in Down syndrome have similar or unique attributes.”
The study also leads to some tantalizing questions. First, since cellular aging is a stress response, the new findings raise questions on the mechanistic underpinnings of the nature of trisomy 21-induced stress. And second, they beg the question whether the increased risk of Alzheimer’s disease in individuals with Down syndrome later in life can be explained by the fact that one of the main genes implicated in Alzheimer’s disease (APP) is located on chromosome 21 or whether the increase in senescence in trisomy 21 compounds the risk.
The study was funded by the Alana Foundation, the LuMind Foundation, Burroughs Wellcome Fund, UNCF-Merck, and the National Institutes of Health.



