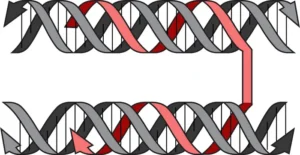
Natural DNA is often double-stranded. Yet, there exists another, lesser-known class of interactions between DNA. These so-called normal or reverse Hoogsteen interactions allow a third strand to join in, forming a beautiful triple helix.
Researchers from the Gothelf lab at Aarhus University have described a general method to organize double-stranded DNA, based on Hoogsteen interactions. Their paper “Folding Double-Stranded DNA into Designed Shapes with Triplex-Forming Oligonucleotides” appears in Advanced Materials.
“The compaction and organization of genomic DNA is a central mechanism in eukaryotic cells, but engineered architectural control over double-stranded DNA (dsDNA) is notably challenging. Here, long dsDNA templates are folded into designed shapes via triplex-mediated self-assembly. Triplex-forming oligonucleotides (TFOs) bind purines in dsDNA via normal or reverse Hoogsteen interactions,” write the investigators.
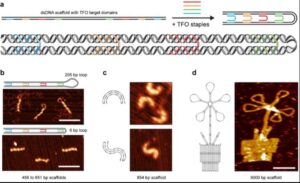
“In the triplex origami methodology, these non-canonical interactions are programmed to compact dsDNA (linear or plasmid) into well-defined objects, which demonstrate a variety of structural features: hollow and raster-filled, single- and multi-layered, with custom curvatures and geometries, and featuring lattice-free, square-, or honeycomb-pleated internal arrangements. Surprisingly, the length of integrated and free-standing dsDNA loops can be modulated with near-perfect efficiency; from hundreds down to only six bp (2 nm).
“The inherent rigidity of dsDNA promotes structural robustness and non-periodic structures of almost 25.000 nt are therefore formed with fewer unique starting materials, compared to other DNA-based self-assembly methods. Densely triplexed structures also resist degradation by DNase I. Triplex-mediated dsDNA folding is methodologically straightforward and orthogonal to Watson-Crick-based methods. Moreover, it enables unprecedented spatial control over dsDNA templates.”
The study demonstrates that triplex-forming strands are capable of sharply bending or “folding” double-stranded DNA to create compacted structures. The appearance of these structures ranges from hollow two-dimensional shapes to dense 3D constructs and everything in-between, including a structure resembling a potted flower. Gothelf and co-workers have named their method triplex origami.
With triplex origami, scientists can achieve a level of artificial control over the shape of double-stranded DNA that was previously unimaginable, thereby opening new avenues of exploration, according to the Aarhus University researchers. It has recently been suggested that triplex formation plays a role in the natural compaction of genetic DNA and the current study may offer insight into this fundamental biological process.
Potential in gene therapy and beyond
The work also demonstrates that the Hoogsteen-mediated triplex formation shields the DNA against enzymatic degradation. Thus, the ability to compact and protect DNA with the triplex origami method may have large implications for gene therapy, wherein diseased cells are repaired by encoding a function that they are missing into a deliverable piece of double-stranded DNA.
Work on DNA sequence and structure has also been applied in nanoscale materials engineering, yielding applications in therapeutics, diagnostics, and many other areas.
“For the past four decades, DNA nanotechnology has almost exclusively relied on Watson-Crick base interactions to pair up single DNA strands and organize them into custom nanostructures, Kurt V. Gothelf, PhD, who is based at the university’s department of chemistry Interdisciplinary Nanoscience Center. “We now know that Hoogsteen interactions have the same potential to organize double-stranded DNA, which presents a significant conceptual expansion for the field.”
Gothelf and co-workers demonstrated that Hoogsteen-mediated folding is compatible with state-of-the-art Watson-Crick-based methods. Due to the comparative rigidity of double-stranded DNA, however, triplex origami structures require fewer starting materials. This allows larger structures to be formed at significantly lower cost.
The novel technique has the limitation that triplex formation typically requires long stretches of purine bases within the double-stranded DNA and, as a result, the researchers have used artificial DNA sequences, instead of natural genetic DNA. However, in the future they will work towards overcoming this limitation.


