
When the sober-minded describe spatial biology, they use words such as fledgling, growing, and maybe even exciting. But perhaps more enthusiasm—and words such as revolutionary and transformative—are in order. Yes, the basic idea behind spatial biology has been around for decades. Put simply, this is the idea that single-cell analysis should reflect how cells are organized in tissues. As familiar as this idea may seem, the toolkit to explore tissues spatially has been limited. But new technologies are making spatial biology a reality, ushering the field into a whole new era. Doesn’t that justify words that might otherwise seem hyperbolic?
To convey the passion surrounding spatial biology, GEN hosted a panel discussion titled, “How Spatial Biology is Revolutionizing Single-Cell Genomics.” It originally aired on October 18 as an episode of GEN Live, our science talk show. And it is still available to anyone who wants to hear about the current status of spatial biology, the impact the technology is making on research (and perhaps one day, patients), and the challenges it faces as it moves forward.

GEN revisits the panel discussion here. Moreover, in sympathy with the basic idea behind spatial biology—that is, the idea of contextualization—we provide some additional background and commentary. We include direct quotes from the two panelists, namely, Rong Fan, PhD, professor of biomedical engineering at Yale University, and Nicholas Banovich, PhD, associate professor in the Integrated Cancer Genomics Division at the Translational Genomics Research Institute (TGen). Our aim is to highlight ongoing trends, challenges that are being overcome, and future possibilities that inspire people to drive spatial biology forward.
Then and now
The discussion kicked off with a broad view of spatial biology’s evolution. To understand how the technology arrived at where it is today, Fan started with a historical perspective, going back to the work of Robert Hooke (in the 1600s) and his early descriptions of how cells appeared when viewed through the microscope. Then, Fan moved to more modern localization techniques used to investigate proteins and nucleic acids spatially in tissues, including immunostaining and fluorescence in situ hybridization. Now, with the newest wave of spatial technologies, researchers have the ability to dig into mechanisms and function.
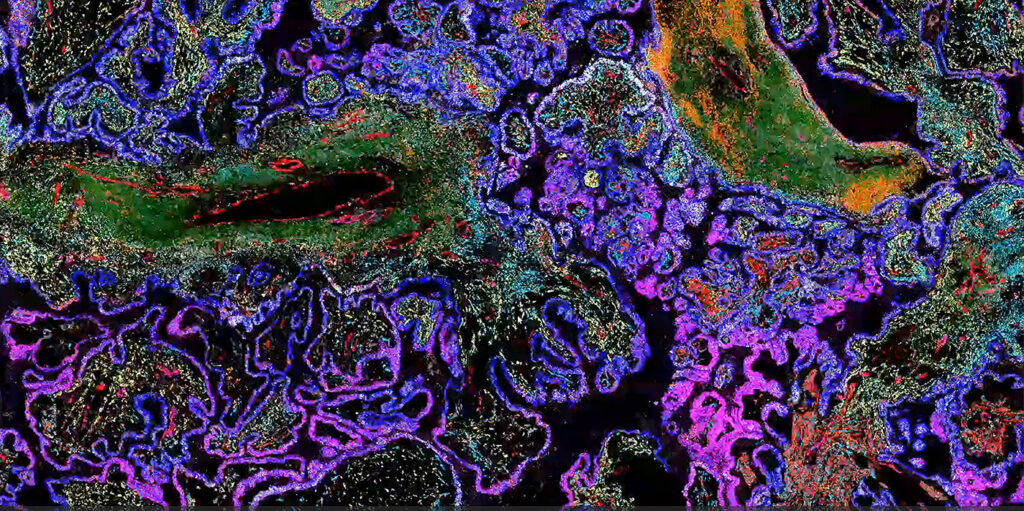
Fan noted that analyzing gene expression is currently the centerpiece of spatial. One reason for that, Banovich noted, is the digitization of the signal. The analysis of data in a way that uses the existing genomic tool set seems to be what allowed spatial biology—particularly spatial transcriptomics—to become broadly useful.
“Whether you’re talking about imaging-based platforms or sequencing-based platforms, being able to churn these through the genomics infrastructure has really led to this broad adoption of the technologies,” Banovich remarked. And because the transcriptomics signal is straightforward to digitize—compared to proteomics or epigenomics signals—it was the first to emerge as the dominant technology.
Fan added that there will be more access to investigate above (proteins and metabolites) and below (the genome and epigenome) the RNA in the cell. And that will generate a new question: Can we link all of them together?
Where are we now?
“This feels to me very much like the single-cell world of 2016 or 2017,” Banovich said, “where everybody is ‘out in the woods,’ trying to figure out exactly what they can eke out of these technologies.” He explained that there are a lot of researchers wrapping their heads around what they can do with these data. Getting relational information, Banovich continued, is the gold mine of spatial data. But there is a lot of work going into understanding how to use the relational information, figuring out what tools are available to answer those questions, and designing the computational tools that still need to be built.
In the early days of single-cell biology, the sharing of methods and experiences was commonplace. And Banovich expressed his hope that this kind of sharing would be the norm in spatial biology, too. One major difference, he observed, is that single-cell biology was heavily dominated by a single technology. As a result, single-cell biology was given to uniformity. In contrast, there is a huge diversity of spatial technologies, even with respect to a single modality such as transcriptomics.
What they are constantly thinking about in Fan’s laboratory is not just how to standardize, but how to integrate data from different modalities. And even when the data from different modalities is there, he said, there will still be a lot of work in understanding how to put them together. But people are moving in this direction now. He noted that computational biologists are playing a key role in this aspect of spatial.
Collaboration is key
Given the diversity in technologies, there is an increased need among researchers to share openly, making community driven initiatives incredibly valuable. One community that has developed over the past few months is the Global Alliance for Spatial Technologies (GESTALT). It was started by Ioannis Vlachos, PhD, co-director of the Bioinformatics Program, Cancer Research Institute, Beth Israel Deaconess Medical Center; Luciano Martelloto, PhD, associate professor at the University of Adelaide; and Jasmine Plummer, PhD, associate member, St. Jude Faculty and director, Center for Spatial OMICs.
GESTALT holds meetings online that allow spatial users from around the world to come together, share knowledge and challenges, and collaborate. Over 100 people attended the first online meeting. The founders hope that it will be a catalyst in advancing spatial technologies. The focus is on collaboration, knowledge sharing, and innovation that will serve to amplify the impact of spatial technologies across sectors.
GESTALT is a space to foster productive dialog for everyone, provide valuable feedback for active users, advocate for best practices for presenting data, and pool growing expertise to address challenges and foster innovations. The group plans to establish working groups to push the needle on specific topics such as standards and best practices, assay optimization, and data analysis.
The spirit of open science is a terrific part of the single-cell community, Fan stressed. And because the single-cell community comprises many of the researchers moving to spatial, that collaborative spirit will be brought into the spatial world as well.
Bringing it all together
One thing that the spatial community would benefit from is a reference. Fan suggested that the spatial community might follow the example set by the human genome project. Once a reference genome was built, researchers could map everything to the reference genome. In the spatial community, an analogous effort would help researchers make sense of observations and data.
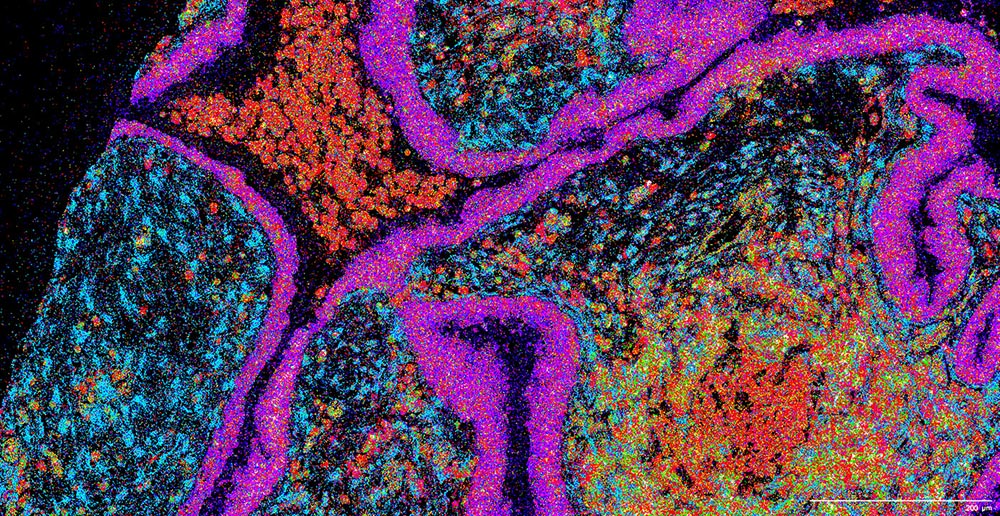
Banovich indicated that he appreciates the efforts of the atlas community over the past couple of years. These efforts include the building and unification of cell atlases across different organs. Once a certain point is reached, researchers could map spatial data to single-cell reference data, unifying datasets and minimizing batch effects. But that, he admitted, poses a huge challenge.
Moving from the collection of spatial data to the generation of insights and knowledge is a long way to go, Fan said. And practical challenges arise from spatial biology’s multidisciplinary nature. To illustrate this point, Fan offered the following example: A pathologist who has decades of experience in tissue embedding knows how to process tissue specimens but is unlikely to know the omics side of the technology. This intrinsic challenge requires people to work together. If people want to succeed, Fan maintained, they need people with extremely diverse backgrounds working together to overcome those barriers.
Magic wand
If given a magic wand, what would Fan and Banovich wish for to move spatial forward? An increase in plex, sensitivity, or resolution? Multiomics in one run? Or more sophisticated bioinformatics?
Banovich answered, laughing, that if the wand is really magic, he would change them all! The reason it is so hard to pick one, he explained, is because it is too early to know which of these things will really elevate spatial to the next level. He also contributed this brief self-dialogue: “Would I love an increase in plex? Absolutely. Does that come with a hit to sensitivity? Probably.”
He said that he would be incredibly excited about a 10,000-plex spatial transcriptomics maps. But he added that if such maps were to come with a 75% sensitivity drop, he would immediately feel less excited. A 10,000-plex capability today, with no drop in sensitivity, would change the game for the spatial platforms.
Fan told GEN that he “wants everything, for sure.” And that is not too far out of range; it is becoming possible. He emphasized that the field should embrace artificial intelligence and machine learning for computational data. The real revolution of the field, he asserted, will come when computer scientists, and artificial intelligence researchers, join the force to write the next chapter.



