As powerful as genome editing using CRISPR-Cas is, there are limitations to the technology. One of those has been the ability to determine genotypes of individual CRISPR-Cas transfected cells. Now, a research group from the Max Planck Florida Institute for Neuroscience (MPFI) has developed a strategy for single-cell genotyping in CRISPR-Cas transfected neurons and showed that they could identify the genetic cause of phenotypes in vivo.
The work is published in “In Vivo Single-Cell Genotyping of Mouse Cortical Neurons Transfected with CRISPR/Cas9” in Cell Reports.
“CRISPR/Cas9-based gene targeting holds great promise for systematic understanding of the molecular basis underlying the assembly, function, and dysfunction of neural circuits,” notes Hiroki Taniguchi, Ph.D., Research Group Leader of Development and Function of Inhibitory Neural Circuits Group at the MPFI. “The perfect matching between genotypes determined by our single cell sequencing and those deduced from phenotype evaluation, suggests that our approach is a powerful new method capable of enhancing the reliability and expanding the applications of CRISPR-based techniques.”
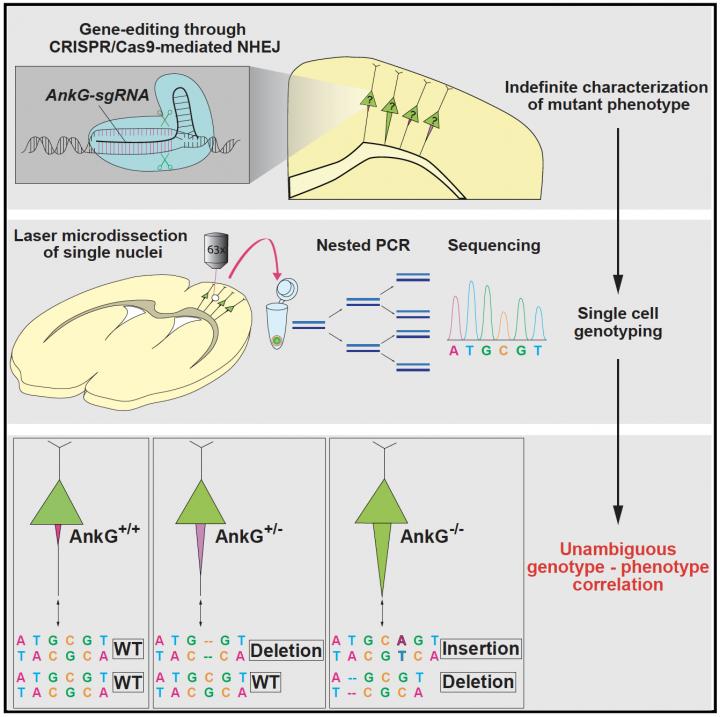
The authors explain that non-homologous end joining (NHEJ) “randomly creates different types of mutations such as deletions, substitutions, and insertions, which can result in hypomorph, loss-of-function, and gain-of-function alleles.” On top of that, the mutations in CRISPR-Cas transfected cells could occur in a mono-allelic or bi-allelic manner.
“Though CRISPR precisely targets a gene of interest, due to NHEJ, its effects can be highly variable,” explains Andre Steinecke, Ph.D., Research Fellow at MPFI and first author of the publication. “CRISPR can leave cells with either fully nonfunctional genes, weakened genes or sometimes even enhance their function.” Steinecke asserts that “this isn’t such a problem when removing one that causes a very noticeable effect in cells because you can easily visualize the change and absence of the protein coded by the gene. But some, especially genes in the brain, don’t have strikingly obvious effects or are very difficult to visualize.”
The team’s goal was to create a widely applicable strategy, capable of reliably determining the exact genetic cause and correlate it to observed phenotype. To do this, they combined laser microdissection with single cell genotyping to design an experimental pipeline capable of studying CRISPR mediated effects in cells while accurately ascertaining the exact DNA changes that caused them.
The researchers show that re-sectioning of cortical slices and subsequent laser microdissection allowed them to isolate individual CRISPR-Cas transfected neurons. Sequencing of PCR products containing a CRISPR-Cas targeted genomic region in single reference neurons provided genotypes that completely correspond with those deduced from their target protein expression and phenotypes, establishing a powerful strategy to determine the causality between genotypes and phenotypes in CRISPR-Cas transfected neurons.
To validate their strategy, the team at MPFI designed CRISPR technology to target a gene in pyramidal neurons encoding a critical structural protein, called Ankyrin-G (AnkG). Normally, the AnkG protein is confined to a specialized region of the neuron known as the axon initial segment (AIS), which is responsible for generating action potentials. When AnkG is removed, the AIS undergoes a noticeable thickening that can be detected using microscopy. With this characteristic feature, neurons that lack AnkG could be readily distinguished and their exact genotype could then be confirmed. They found that predominately, neurons transfected with their CRISPR probe exhibited a loss of AnkG as well as substantially thickened AIS. But a small portion of neurons transfected with CRISPR still exhibited AnkG levels and AIS thickness comparable with wildtype neurons; demonstrating the varying effects of CRISPR on different cells.
To probe and confirm the underlying genetic causes, the team then used laser microdissection to isolate and extract individual neurons whose phenotype had already been characterized. Once extracted, the team sequenced each individual cell separately to determine the genotype. They found that their strategy could reliably and reproducibly link observed phenotype to genotype, where neurons lacking AnkG with thickened axons showed loss-of-function mutations in both copies of the gene whereas neurons with normal levels of AnkG either showed mutations in only one copy (neurons transfected with CRISPR) or normal genotypes (control neurons). The team then confirmed their strategy using two additional genes, MeCP2 and Satb2, finding that their process could once again effectively correlate observed feature to underlying genetics. This novel protocol will open up new avenues of study for neurobiology and further upgrade the already powerful abilities of CRISPR.