A deep understanding of the complex and crowded cellular proteome would emerge if individual proteins could be sequenced, quantified, and screened for the presence of modifiers.
Using an integrated semiconductor device and their trademarked time domain sequencing technology, Quantum Si, a life sciences company commercializing proteomics solutions, has developed a next-generation, real-time, single-molecule protein sequencing technology. Details of the method were published in the journal Science on October 13, 2022. The novel technology provides a more comprehensive way to study proteins and could transform drug discovery and diagnostics by elevating protein sequencing to the current convenience of genome sequencing.

Senior author of the paper, Jonathan Rothberg, PhD, founder and chairman of Quantum-Si said, “I am proud and excited to share our first next-generation single-molecule protein sequencing technology with the world. Just as the work I did with my first genomics collaborators on DNA sequencing led to a Nobel Prize for Svante Pääbo, I fully expect the early adopters of Quantum-Si’s technology to make discoveries just as profound and impactful.”

Lead and corresponding author of the study, Brian Reed, PhD, head of research at Quantum-Si said, “The concept of a small benchtop instrument carrying out massively-parallel sequencing of individual protein molecules was previously unimaginable. Researchers have used indirect methods to understand proteins and have been waiting for tools that would be transformative for proteomics in the same way that DNA sequencing has been for genomics.”
The integrated semiconductor device used in Quantum-Si’s protein sequencing technology contains millions of wells. This enables the cataloging of many proteins in parallel and the detection of post-translational modifications. The single-molecule protein sequencing technology marks single peptides in real time by a mixture of dye-labeled residues at the amino terminal that the authors call “recognizers”, and simultaneously cleaves them using aminopeptidase enzymes. Quantification of fluorescence intensity, longevity, and binding characteristics of the amino-acid labels on the semiconductor chip, then leads to the annotation of each amino acid residue and identification of a peptide’s sequence.
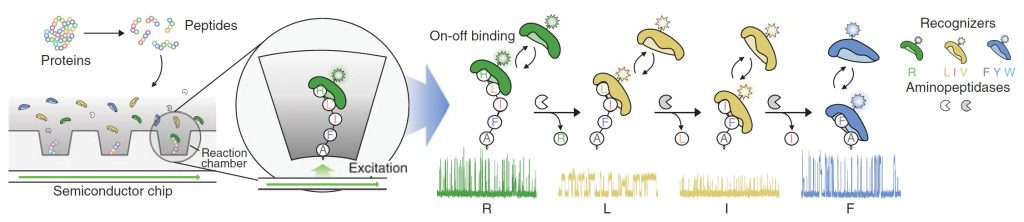
Reed said, ”Our platform’s ability to map protein modifications, which have been difficult to detect with other technologies, will greatly advance our understanding of the function and regulation of proteins in health and disease.”
“The authors report an interesting approach to sequencing peptides based on optical detection of proteins that recognize N-terminal amino acids,” said Nikolai Slavov, PhD, director of the single-cell proteomics center at the Northeastern University College of Engineering. “This proof of principle study outlines obvious next steps and leaves open the question whether it can scale to the complexity of eukaryotic proteomes, both in terms of specificity and counting throughput.” (Slavov was not involved in the study).
In future studies the investigators hope to further develop the technology to offer a sensitive, scalable, and accessible platform for single-molecule protein sequencing.