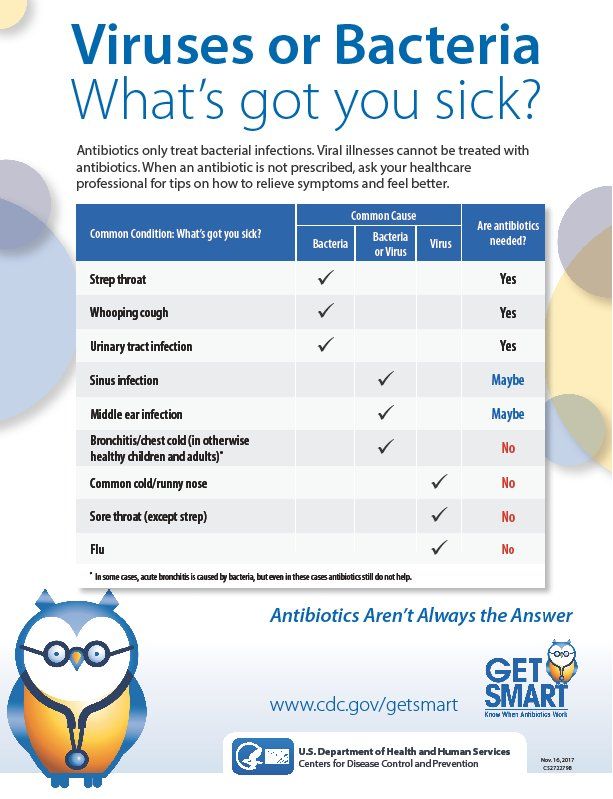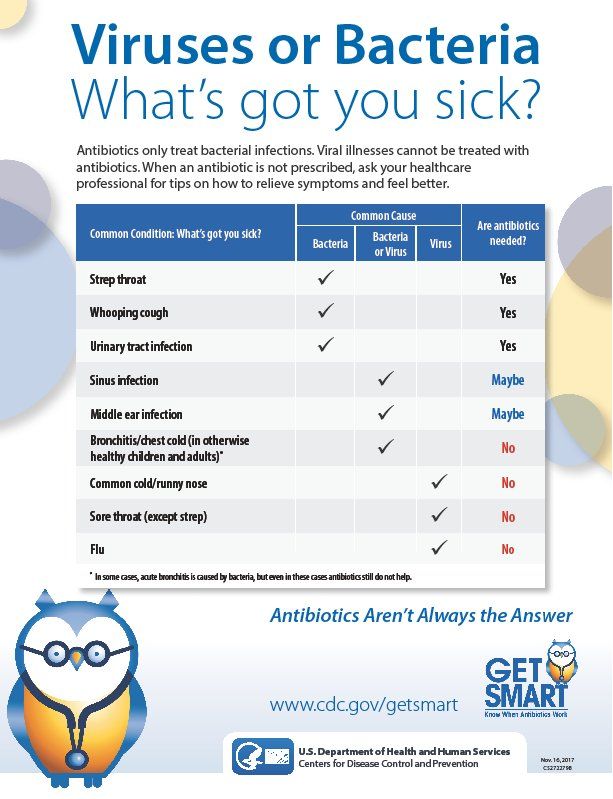
The ability of clinicians to discriminate between bacterial and viral infections among their patients is critical, not only to administer the appropriate therapeutic intervention but to help quell the rise in a major global health threat—antibiotic resistance stemming from overuse. Unfortunately, ailments such as lower respiratory tract infections (LRTIs) symptomatically present with similar clinical symptoms, regardless of the root pathogen—making proper diagnosis difficult.
Now, investigators at the University of Rochester Medical Center (URMC) have released findings from a new study that utilized the power of RNA sequencing (RNA-Seq) and transcriptomic analysis to help physicians prescribe antibiotics to patients who need them and avoid giving them to individuals who don't. Results from the new study were published recently in Scientific Reports in an article entitled “Transcriptomic Biomarkers to Discriminate Bacterial from Nonbacterial Infection in Adults Hospitalized with Respiratory Illness.”
“It's extremely difficult to interpret what's causing a respiratory tract infection, especially in very ill patients who come to the hospital with a high fever, cough, shortness of breath, and other concerning symptoms,” noted senior study investigator Ann Falsey, M.D., professor and interim chief of the infectious diseases division at URMC’s Strong Memorial Hospital. “My goal is to develop a tool that physicians can use to rule out a bacterial infection with enough certainty that they are comfortable, and their patients are comfortable, foregoing an antibiotic.”
The research team enlisted 94 adults hospitalized with LRTIs, gathered clinical data, took blood from each patient, and conducted a battery of microbiologic tests to determine which individuals had a bacterial infection (41 patients) and which had a nonbacterial or viral infection (53 patients). Moreover, the scientists used a complex genetic and statistical analysis to pinpoint markers in the blood that correctly classified the patients with bacterial infections 80% to 90% of the time.
“We evaluated whole blood profiling using RNASeq to discriminate infectious agents in adults with microbiologically defined LRTI,” the authors wrote. “Hospitalized adults with LRTI symptoms were recruited. Clinical data and blood were collected, and comprehensive microbiologic testing performed. Gene expression was measured using RNASeq and qPCR [quantitative PCR]. Genes discriminatory for bacterial infection were identified using the Bonferroni-corrected Wilcoxon test. Constrained logistic models to predict bacterial infection were fit using screened LASSO. We enrolled 94 subjects who were microbiologically classified; 53 as “non-bacterial” and 41 as “bacterial.” RNAseq and qPCR confirmed significant differences in mean expression for 10 genes previously identified as discriminatory for bacterial LRTI.”
According to the Centers for Disease Control and Prevention, antibiotic-resistant bacteria cause at least 2 million infections and 23,000 deaths each year in the United States. The use of antibiotics is the single most important factor leading to antibiotic resistance around the world, making this type of research more important than ever.
“Our genes react differently to a virus than they do to bacteria,” remarked co-senior study investigator Thomas Mariani, Ph.D., professor of pediatrics and biomedical genetics at URMC. “Rather than trying to detect the specific organism that's making an individual sick, we're using genetic data to help us determine what's affecting the patient and when an antibiotic is appropriate or not.”
While the researchers were excited by their findings and optimistic about the use of the technology to better diagnose patients, they did point out that the main limitation of their study is the small sample size and that the genetic classifiers selected from the study population may not prove to be universal to all patients.