SNIPRs, easy-to-build molecules that can detect point mutations in RNA, have been developed by scientists at Arizona State University (ASU). The scientists have demonstrated that SNIPRs, or Single-Nucleotide-Specific Programmable Riboregulators, operate in live cells and in paper-based cell-free reactions. Because they can be designed to trigger colorimetric reactions, SNIPRs may simplify applications such as human genotyping, virus detection, and viral strain identification. In fact, SNIPRs may prove to be especially useful in the developing world.
To date, point mutations have been hard to detect, even in places where sophisticated testing facilities are readily available, much less in regions where they’re scarce. Yet point mutations, which are genomic errors involving a single base in a length of DNA or RNA, are of intense interest everywhere.
Point mutations can result in mild abnormalities, such as color blindness, as well as serious diseases, such as neurofibromatosis, sickle-cell anemia, certain forms of cancer, and Tay-Sachs disease. Mutations can also produce disease variants that are resistant to conventional treatment.
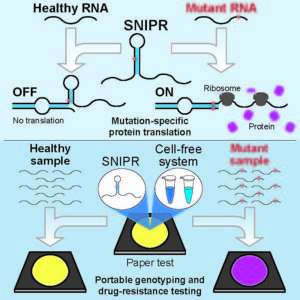
To simplify the detection of point mutations, the ASU team, led by Alexander A. Green, PhD, assistant professor, Biodesign Center for Molecular Design and Biomimetics, based their SNIPR system on prokaryotic translational riboregulators. Within a cell, these riboregulators activate when they encounter a target RNA sequence, that is an RNA sequence containing a point mutation, which typically corresponds to a point mutation in one of the cell’s genes.
The riboregulators rely on conformational changes that occur upon binding of a target RNA to activate protein production. Prior to target binding, a riboregulator adopts an OFF-state configuration. Binding of the target RNA triggers a transition to the translationally active ON-state. If the binding of a cell’s mutant RNA with the trigger strand is exact, the SNIPR unfolds, allowing sequence access by the ribosome—the machinery required to translate RNA into protein. If, however, the SNIPR encounters an unmutated sequence, there is a mismatch and translation of protein is blocked.
This functionality was achieved in research conducted by the ASU scientists and reported February 27 in the journal Cell, in an article titled, “Precise and Programmable Detection of Mutations Using Ultraspecific Riboregulators.”
“SNIPRs provide over 100-fold differences in gene expression in response to target RNAs differing by a single nucleotide in Escherichia coli and resolve single epitranscriptomic marks in vitro,” the article’s authors wrote. “By exploiting the programmable SNIPR design, we implement an automated design algorithm to develop riboregulators for a range of mutations associated with cancer, drug resistance, and genetic disorders.”
The article explained that SNIPRs can detect subtle differences in so-called binding or hybridization energy. “Typically, when you’re thinking about a DNA or RNA base pairing, it’s through hydrogen bonds,” Green noted. “When G binds to C, that’s three hydrogen bonds. And when A binds to U, that’s two hydrogen bonds.” In addition to point mutations, in vitro analysis can detect minor differences in binding energy when epigenetic changes like methylation occur.
The article’s authors assert that their method offers a rapid, highly accurate, and inexpensive means of identifying mutations relevant to human health. The method can be used in conjunction with paper-based tests and produce color-based displays through reactions powered by human body heat.
“Advances in the method could one day be used as a low-cost alternative for personal genotyping,” added Hao Yan, PhD, a co-author of the new study and director of ASU’s Biodesign Center for Molecular Design and Biomimetics. “The simplicity of the technique may allow at-home screening for disease-linked mutations, providing rapid and accurate testing, while maintaining data privacy for users.”
In addition to its convenience as an inexpensive, versatile litmus test for mutation-related illness, the technique promises to shed new light on foundational issues in cell biology, including genetic resistance to antibiotics and mutations leading to the failure of frontline treatments for diseases like malaria and HIV.
First author Fan Hong, formerly with the Biodesign Institute and now a postdoctoral fellow at Harvard, designed computer algorithms that allow for the efficient design of SNIPRs based on desired RNA target sequences. “To make SNIPRs easy to use, we automated the process so that everybody can design them without any knowledge of RNA folding and RNA interactions,” Hong said. “They already show lots of practical applications such as human genotyping, Zika virus detection, and viral strain identification.”
Identifying particular strains is of vital importance epidemiologically. Some genetic variants of Zika, for example, appear to pose a greater risk of birth abnormalities, while the currently circulating coronavirus is also evolving and has a very similar sequence to the coronavirus that caused the SARS epidemic in 2002–2003.
Certain point mutations in HIV can lead to the failure of common antiretroviral therapies. A SNIPR test for such mutations could rapidly identify these mutations and guide appropriate treatment. Conventional tests for HIV drug resistance are prohibitively expensive for many in need, costing over $200 dollars per sample.
The method could also offer new hope in the fight against cancer. For example, granulosa cell tumors result from just a single incorrect base out of the three billion nucleotide pairs that make up the genetic code, while the point mutations in tumor-suppressing BRCA1 and BRCA2 genes are responsible for a six-fold increase in the lifetime risk for breast cancer.
The fine-grained sensitivity of SNIPRs can discriminate between patients who are heterozygous or homozygous for given mutations, that is, whether they carry one or two copies of the mutated gene on their chromosomes, a critical factor in determining disease vulnerability.
When SNIPR probes are combined with paper-based recognition systems, the potential for rapid, low cost, and precise detection of genetic point mutations can be extended globally, wherever such diagnostic tools are most critically needed. Additionally, SNIPRs promise to help researchers understand strain variations and mutation-linked resistance to common therapeutics.


