DNA carries the blueprint for building bodies, but it’s a living document: Adjustments to the design can be made by epigenetic marks. In humans and other eukaryotes, two principal epigenetic marks are known. A team from the Marine Biological Laboratory (MBL) has now discovered a third epigenetic mark—which had previously only been known only in bacteria—in tiny freshwater invertebrates known as bdelloid rotifers. This is the first time that a horizontally transferred gene has been shown to reshape the gene regulatory system in a eukaryote.
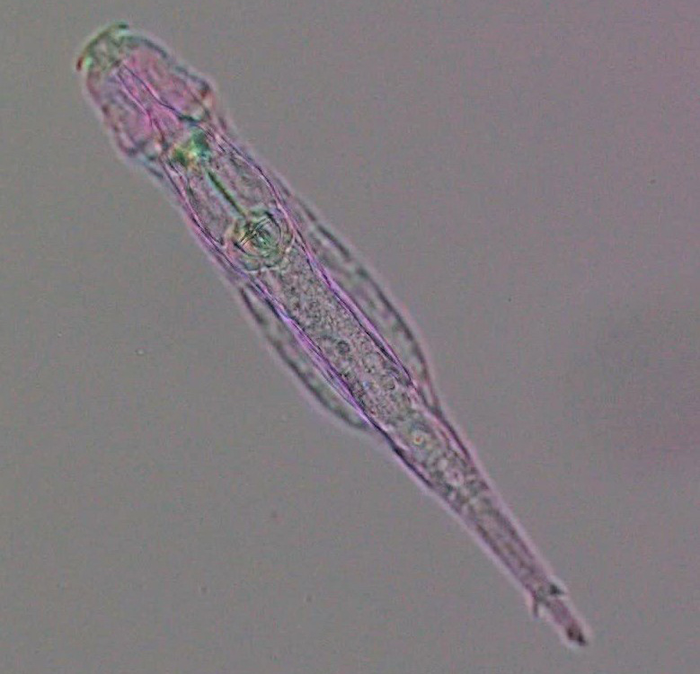
A feeding bdelloid rotifer (Adineta vaga) under polychromatic polarization microscope. [M. Shribak and I Yushenova]
“We discovered back in 2008 that bdelloid rotifers are very good at capturing foreign genes,” said research lead Irina Arkhipova, PhD, a senior scientist at MBL’s Josephine Bay Paul Center. “What we’ve found here is that rotifers, about 60 million years ago, accidentally captured a bacterial gene that allowed them to introduce a new epigenetic mark that was not there before.”
It’s too early to know what the implications may be of discovering this new epigenetic system in rotifers. “A good comparison is the CRISPR-Cas system in bacteria, which started out as a basic research discovery,” said Fernando Rodriguez, PhD, a research scientist in the Arkhipova lab, and co-first author of the team’s published paper in Nature Communications. “Now CRISPR-Cas9 is used everywhere as a tool for gene editing in other organisms. This is a new system. Will it have applications, implications for future research? It’s hard to tell.”
Rodriguez is co-first author of the scientists’ published study, which is titled, “Bacterial N4-methylcytosine as an epigenetic mark in eukaryotic DNA,” in which they noted, “we combine multiple lines of evidence to establish that 4mC modification can be recruited as an epigenetic mark in eukaryotic genomes, and to characterize the underlying enzymatic machinery … Our work demonstrates how a horizontally transferred gene can become part of a complex regulatory system maintained by selection over tens of millions of years of evolution.”
Epigenetic marks are modifications to DNA bases that don’t change the underlying genetic code, but “write” extra information on top of it that can be inherited with the genome. In the two already known epigenetic marks in eukaryotes, a methyl group is added to a DNA base, either cytosine or adenine. Epigenetic marks usually regulate gene expression—they turn genes on or off—particularly during early development or when your body is under stress. They can also suppress “jumping genes,” which are transposable elements that threaten the integrity of the genome.
“In eukaryotes, modifications predominantly involve C5-methylcytosine (5mC) and occasionally N6-methyladenine (6mA), while bacteria frequently use N4-methylcytosine (4mC) in addition to 5mC and 6 mA,” the authors wrote. Eukaryotes mostly use base modifications for regulatory purposes, with 5mC representing the predominant form of epigenetic modification in eukaryotic genomes. “Often called ‘the fifth base,’ 5mC plays an important role in genome defense against mobile genetic elements, and is often associated with transcriptional silencing, establishment of the closed chromatin configuration, and repressive histone modifications,” the team added.
4mC has not been demonstrated to act as an epigenetic mark in eukaryotes, the scientists stated, “ … and most claims of eukaryotic 4mC lack confirmation by orthogonal methods and do not identify the enzymatic component. In fact, 4mC is also a cytosine modification, but with a distinct bacterial-like positioning of the methyl group—essentially recapitulating evolutionary events of over two billion years ago, when the conventional epigenetic marks in early eukaryotes emerged.
Bdelloid rotifers are extremely resilient animals, as the Arkhipova and David Mark Welch labs at MBL have discovered over the years. The organisms can dry up (desiccate) completely for weeks or months at a time, and then spring back to life when water becomes available. During their desiccation phases, the bdelloid rotifer DNA breaks up into many pieces. “When they rehydrate or otherwise render their DNA ends accessible, this might be an opportunity for foreign DNA fragments from ingested bacteria, fungi, or microalgae to transfer into the rotifer genome,” Arkhipova said. About 10% of the rotifer genome comes from non-metazoan sources, they have found.
Still, the Arkhipova lab was surprised to find a gene in the rotifer genome that resembled a bacterial methyltransferase (a methyltransferase catalyzes the transfer of a methyl group to DNA). “We hypothesized that this gene conferred this new function of suppressing transposons, and we spent the last six years proving that, indeed, it does,” Arkhipova said. As the authors commented, “We identified N4CMT, a horizontally transferred enzyme of bacterial origin, as responsible for the addition of 4mC marks to DNA … “Our results show how non-native DNA methyl groups can reshape epigenetic systems to silence transposons and demonstrate the potential of horizontal gene transfer to drive regulatory innovation in eukaryotes,” the investigators stated in their paper.
This is very unusual and has not been previously reported,” Arkhipova added. “Horizontally transferred genes are thought to preferentially be operational genes, not regulatory genes. It is hard to imagine how a single, horizontally transferred gene would form a new regulatory system because the existing regulatory systems are already very complicated.”
“It’s almost unbelievable,” said co-first author Irina Yushenova, PhD, a research scientist in Arkhipova’s lab. “Just try to picture, somewhere back in time, a piece of bacterial DNA happened to be fused to a piece of eukaryotic DNA. Both of them became joined in the rotifer’s genome and they formed a functional enzyme. That’s not so easy to do, even in the lab, and it happened naturally. And then this composite enzyme created this amazing regulatory system, and bdelloid rotifers were able to start using it to control all these jumping transposons. It’s like magic.”
“You don’t want transposons jumping around in your genome,” said Rodriguez. “They will mess things up, so you want to keep them in check. And the epigenetic system to accomplish that is different in different animals. In this case, a horizontal gene transfer from bacteria into bdelloid rotifers created a new epigenetic system in animals that hasn’t been described before.”
“Bdelloid rotifers, especially, have to keep their transposons in check because they primarily reproduce asexually,” Arkhipova noted. “Asexual lineages have fewer means for suppressing proliferation of deleterious transposons, so adding an extra layer of protection could prevent a mutational meltdown. Indeed, transposon content is much lower in bdelloids than it is in sexual eukaryotes that don’t have this extra epigenetic layer in their genome defense system.”
The new discoveries could open the door to the development of new tools and research directions for investigating genome function and resilience in this rotifer system. And as the authors concluded, “Collectively, our findings help to unravel a fascinating evolutionary puzzle: How can a bacterial enzyme decorating DNA with non-metazoan modifications penetrate eukaryotic gene silencing systems and become preserved by natural selection for tens of millions of years?”
The system, they added, “… demonstrates that horizontal gene transfer, the role of which in eukaryotic regulatory evolution is a subject of intense debate, can re-shape complex regulatory circuits in metazoans, thereby driving major evolutionary innovations that include epigenetic control systems.”