The need for a more complete understanding of primate genetic diversity is urgent, especially given the threats posed by factors such as climate change, habitat loss, and illegal trading and hunting. Characterizing primate variation not only allows us to better understand and conserve these species in the wild, but it also helps us to better understand ourselves.
“Humans are primates,” noted Tomàs Marquès-Bonet, PhD, an ICREA researcher at the IBE (CSIC-UPF) and a professor of genetics at the department of medicine and life sciences (MELIS) at Pompeu Fabra University (UPF). “The study of hundreds of nonhuman primate genomes, given their phylogenetic position, is very valuable for human evolutionary studies, to better understand the human genome and the bases of our singularity, including the bases of human diseases, and for their future conservation.”
Despite the importance of nonhuman primates, reference genomes have been sequenced in <10% of species, which both impedes research and hampers conservation efforts. The publication of a new project includes the most complete catalog of primate genomic information produced so far, covering nearly half of all existing primate species on Earth. It contains information on primates from Asia, America, Africa, and Madagascar.
The papers are included in a special issue of Science, including two related studies from Science Advances, totaling ten papers that provide new insights into primate genomes—far exceeding past genomic analyses of primate species.
One study presents high-quality reference genomes for 27 primate species, adding to available resources. Of particular interest, they report a previously unreported increase in the rate of genomic change in the Simiiformes common ancestor that may have played a role in the later diversification of Simiiformes and the evolution of humans.
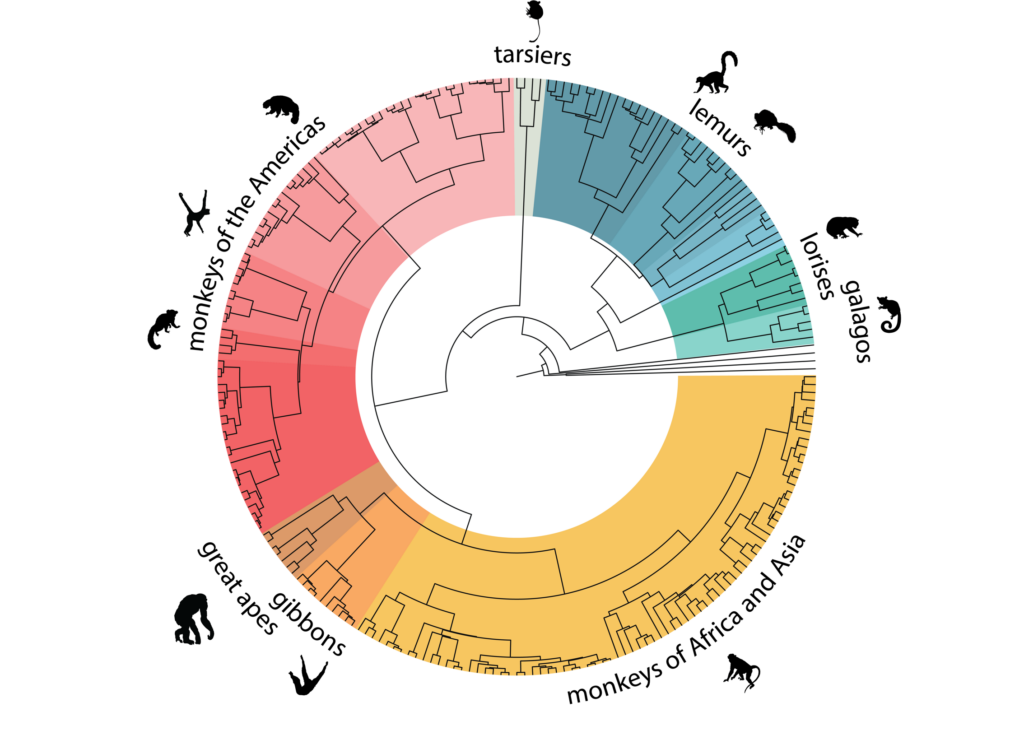
In another study—results from which provide the foundation for several additional studies in the special issue—researchers present whole genome data from 233 primate species representing 86% of the primate genera and all 16 families. The team led by Marquès-Bonet, Kyle Farh, MD, PhD, vp & distinguished scientist, Illumina Artificial Intelligence Lab, and Jeffrey Rogers, PhD, associate professor of molecular and human genetics at Baylor College of Medicine, used this dataset to estimate the human-chimpanzee divergence as between 9.0 and 6.9 Ma—slightly older than other recent analyses. The authors also explored the association between genomic variation and variables including climate and sociality and studied whether genetic diversity estimates are correlated with extinction risk in primates, a subject of previous debate. “Despite our broad sampling,” the authors wrote, “we find no global relationship between numerically coded International Union for Conservation of Nature extinction risk categories and estimated heterozygosity.” Finally, the authors used the data to generate a better picture of the mutations that arose in the human lineage and have not emerged elsewhere in primates.
By comparing the genomes of 809 nonhuman primate individuals from 233 species to the human genome, the research identified 4.3 million common missense mutations that affect the composition of amino acids, leading to many human diseases.
Asian colobines, also known as leaf-eating monkeys, have been on the planet for about 10 million years. Their ancestors crossed land bridges, dispersed across continents, survived the expansion and contraction of ice sheets, and learned to live in tropical, temperate, and colder climes. A study in the set finds parallels between Asian colobines’ social, environmental, and genetic evolution, revealing for the first time that colobines living in colder regions experienced genetic changes and alterations to their ancient social structure that likely enhanced their ability to survive.
Another study, led by Iker Rivas-González, a PhD student in the Bioinformatics Research Center at Aarhus University, addresses questions around the process of speciation among populations, focusing specifically on the way some regions of the genome will not show evidence of difference for a long time after ecological speciation has occurred—a process called incomplete lineage sorting. By accounting for incomplete lineage sorting across primates, the researchers were able to produce a primate phylogeny that agrees with fossil estimates, unlike past attempts.
A set of authors led by Hong Wu, PhD, at Yunnan University, studied the genome sequences from a group of monkey species in the Rhinopithecus genus—focusing on hybridization in mammalian evolution, the role for which is rarely examined—and found evidence that the gray snub-nosed monkey is derived from hybridization between the golden snub-nosed monkey and the ancestor of two extant Rhinopithecus species. Further, they report the unusual coat color seen in the gray snub nose is due to this mixing.
One group of species that has been identified as having a history of hybridization is that containing the genus Papio—the baboons. Erik Sørensen, PhD, professor in organic chemistry at Princeton University, and colleagues used whole genome sequencing to reveal the evolutionary history of overlapping baboon species and found evidence of repeated admixture. “We describe the first example of a baboon population with a genetic composition that is derived from three distinct lineages,” they wrote.
Also focused on hybridization in primates, a report in Science Advances compared reference genomes of 12 macaque species that together cover all the known macaque groups. The comparative phylogenomic analysis uncovered an ancient hybrid origin of a macaque lineage. “Our study provides both a strategy and a pipeline of genome analyses to identify hybrid speciation, which should pave the way for the identification and exploration of further such events in the future,” noted the authors.
Lastly, the new genomic catalog has halved the number of genomic innovations that were believed to be exclusively human. This observation facilitates the identification of mutations that are not shared with primates that may consequently be unique to human evolution and the characteristics that make us human.
“Our studies,” noted Kuderna, “provide clues as to which species are in most dire need of conservation efforts, and could help to identify the most effective strategies to preserve them.”




