Those hoping to uncover new science may feel disheartened if they struggle with inadequate tools and doubt that more capable tools are within reach. Until recently, such was the case in proteomics. Existing tools sufficed for bulk analysis, but not single-cell analysis. Without better tools, a new proteomics—one that could explore cellular heterogeneity, detect rare cells, and recognize fleeting cell states—would forever remain a tantalizing possibility.
It was unclear if the main tool in proteomics, the mass spectrometer, could attain the necessary sensitivity. Worse, there was the sense that even the most sensitive mass spectrometer could be defeated by uncertain workflows, that is, workflows in which tiny samples deteriorated or dwindled to almost nothing before they even reached the instrument. Protein quantities, unlike nucleic acid quantities, cannot be amplified.
Pioneering researchers
Remarkably, despite the challenges of mass spectrometry–based single-cell proteomics (MS-SCP), researchers have remained undaunted. These researchers, as mentioned in previous issues of GEN, include Nikolai Slavov, PhD, director of the Single-Cell Proteomics Center at Northeastern University, and Matthias Mann, PhD, director and scientific member of the Max Planck Institute of Biochemistry.
Slavov and colleagues developed Single Cell ProtEomics by Mass Spectrometry (SCoPE-MS), a method that takes advantage of isobaric mass tags and sample pooling of test and carrier samples. It has been used to quantify proteome heterogeneity during cell differentiation. Also, a more automated version of SCoPE-MS called SCoPE2 has been used to quantify the emergence of macrophage heterogeneity.1
Mann’s team integrated high-content imaging, laser microdissection, and multiplexed MS to develop a technology called single-cell Deep Visual Proteomics (scDVP). It has been used to resolve the context-dependent, spatial proteome of murine hepatocytes at a depth of 1,700 proteins from individual cells.2
In addition to making the most of extant and widely available technologies, researchers can take advantage of emerging technologies—sometimes in the same workflow, as demonstrated by a recent webcast (“New Developments in Mass Spectrometry–Based Single-Cell Proteomics”) featuring Karl Mechtler, PhD, head of the Proteomics Tech Hub at the Vienna BioCenter’s Research Institute of Molecular Pathology.3 He described how he led a workflow project in which the “primary task was to bring together the best components and [achieve] a beautiful symbiosis.”
The project reflected Mechtler’s conviction that all the steps in MS-SCP workflows need to be optimized. In general, these steps include cell isolation, collection, and lysis, protein extraction and digestion; peptide separation; mass spectrometry analysis; data acquisition; and data processing and analysis.
Workflows are variable and customizable. Some steps may be accomplished via different technologies. Here a just a few possibilities: Cell separation may be accomplished via flow cytometry or piezoacoustic technology. Cell lysis may be accomplished physically or chemically. And peptide separation may be accomplished by liquid chromatography or capillary electrophoresis. Also, workflows may or may not involve labeling reactions.
In the webcast, the workflow was optimized to distinguish two similarly sized human cell types, HeLa and HEK, based on their proteomes. Different versions of the workflow accommodated either label-free or tandem mass tag (TMT)-labeled samples.
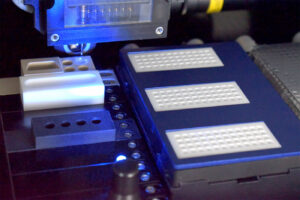
To execute the workflow, Mechtler and colleagues relied on an array of technologies. One of these technologies, the proteoCHIP, was developed by Mechtler’s group in partnership with SCIENION and Cellenion. It’s a nanowell slide for single-cell sample preparation that works with Cellenion’s cellenONE, an instrument that can isolate single cells and dispense them in nanoliter reagent volumes. When a commercial version of the proteoCHIP became available, Cellenion’s founder and managing director, Guilhem Tourniaire, PhD, stated, “This dedicated consumable is going to both facilitate and improve sample preparation for single-cell proteomic studies and will be a cornerstone toward the democratization of this new revolutionary field of biology.”
Other technologies in the workflow included a micropillar-based chromatography column and the Orbitrap Exploris 480 mass spectrometer equipped with the FAIMS Pro Interface (Thermo Fisher Scientific). Ultimately, as Mechtler and colleagues indicated in a bioRxiv preprint,4 the workflow enabled the quantification of “around 1,000 protein groups per analytical run at remarkable reporter ion signal to noise while reducing or eliminating the carrier proteome.” The researchers added, “We are confident that our versatile, sensitive, and automated sample preparation workflow will be easily adoptable by nonspecialized groups and will drive biological applications of single-cell proteomics.”
Standardization advocates
Because MS-SCP workflows can vary and rely on varied technology platforms, there is interest in standards development. Such interest was evident in a recent review by Rushdy Ahmad, PhD, and Bogdan Budnik, PhD, scientists at the Wyss Institute for Biologically Inspired Engineering.5 Although the reviewers stated that “it has now become abundantly clear that [MS-SCP] is a worthy complement to single-cell transcriptomics,” they acknowledged that “there are a number of outstanding and pressing problems that the scientific community vested in advancing this technology needs to resolve.”
General challenges cited by the reviewers include the need for accessible and appropriately priced products, and the need for expertise in cell sorting, specialized cell lysis techniques, labeled or non-labeled quantitative methods, sophisticated liquid chromatography/mass spectrometry instrumentation, and advanced statistical analysis techniques. According to the reviewers, these challenges can be overcome if the research community collaborates with industry to produce “robust, industrial strength, easily accessible, appropriately priced products.”
They also emphasized the need to set standards to broaden the accessibility of MS-SCP technology and to facilitate the verification of novel discoveries. “[We] need to build the foundation for standardization on two primary pillars: standards that can be run to qualify the sample preparation workflow and standards that can be used to qualify the instrument and chromatography capabilities of the various systems that are represented in analytical labs worldwide,” the reviewers noted. “Also of importance are the standards pertaining to the analysis workflow.”
The reviewers suggested that if these challenges were met, MS-SCP would be able to expand beyond specialist academic groups and become “ubiquitously applied to elucidating deep biological insights into the diagnosis and treatment of all diseases that afflict us.” They also cited encouraging developments such as the appearance of standards development suggestions in the scientific literature. Examples include proposals from Slavov’s group6 and researchers led by Fabian Theis, PhD,7 scientific director of biomedical AI at Helmholtz Munich, professor of mathematical modeling of biological systems at the Technical University of Munich, and a consultant at Cellarity.
Technology providers
Many extant technology platforms have been adapted for MS-SCP applications. In many cases, these platforms’ MS-SCP capabilities haven’t been the subject of explicit promotion. However, there are several exceptions, most notably a number of mass spectrometry instruments. One of them is the SCIEX ZenoTOF 7600 system, a quadrupole time-of-flight mass spectrometer that the company launched in 2021. More recently—at last June’s annual meeting of the American Society for Mass Spectrometry—Thermo Fisher Scientific introduced the Orbitrap Astral instrument and Bruker launched the timsTOF Ultra.
SCIEX
When SCIEX announced the ZenoTOF 7600’s release, the company asserted that the instrument delivered up to 20 times the sensitivity of conventional time of flight technology and enabled the routine detection of important low-abundance molecules. SCIEX added that the ZenoTOF 7600 could ensure the extraction and quantification of novel structural information from diverse compound types.

Initially, SCIEX didn’t emphasize the ZenoTOF 7600’s single-cell capabilities. Lately, however, SCIEX has been more direct. For example, the company posted a webinar titled, “Democratizing Single-Cell Proteomics with Microflow Time-of-Flight Mass Spectrometry.”8 The webinar featured Ben Orsburn, PhD, an instructor and principal investigator at Johns Hopkins University School of Medicine. He emphasized two points. First, he mentioned the complications of “nanoflow plumbing.” (He exclaimed, “I hate it, hate it, hate it!”) Second, he discussed the advantages of working with a high intrascan linear dynamic range (ILDR).
According to a study cited by Orsburn, “The [ILDR] refers to the maximum abundance ratio between the most abundant and the least abundant signal observable within a given spectrum.”9 In a separate study by Orsburn and colleagues, the authors stated that a “major historical challenge in protein mass spectrometry is the wide intracellular distribution of protein dynamic range which has been estimated to about seven orders in mammalian cells, which is a stark contrast to mass analyzers which may only have a two-order [ILDR].”10 In the webinar, Orsburn reported that his group achieved an ILDR of nearly four orders when it combined a microflow chromatography system, a peptide-signal-boosting carrier channel, and the ZenoTOF 7600.
“Using a KRASG12C model human-derived cell line, we demonstrate the quantification of over 1,200 proteins per cell with high relative sequence coverage permitting the detection of multiple classes of post-translational modifications in single cells,” Orsburn and colleagues noted in their study. “When cells were treated with a KRASG12C covalent inhibitor, this approach revealed cell-to-cell variability in the impact of the drug, providing insight missed by traditional proteomics.”
Thermo Fisher Scientific
When Thermo Fisher introduced the Orbitrap Astral, the company asserted that the new instrument would help “researchers worldwide to uncover proteins that previously evaded detection and make breakthrough discoveries more efficiently than ever [and] to identify new clinical biomarkers, reveal diseases earlier and develop new interventions for everything from cardiovascular disease to cancer.” The Orbitrap Astral joins the company’s MS-SCP lineup, which also includes the Orbitrap Ascend Tribrid and the Orbitrap Exploris 480 instruments. Thermo Fisher notes that the Orbitrap Astral is for experiments that “require thousands of cells to gain real biological insights,” the Orbitrap Ascend Tribrid is for “research-level single-cell proteomics,” and the Orbitrap Exploris 480 is an “entry-level single-cell proteomics system.”

“The Orbitrap Astral provides not only the necessary high throughput and extended proteome depth, but the crucial quantitative precision and accuracy needed to correctly assess dynamic changes and heterogeneity amongst single cells and cell states,” says Aaron Robitaille, PhD, director of marketing, Mass Spectrometry, Thermo Fisher Scientific. “All current generation Orbitrap-based mass spectrometers equipped with the FAIMS interface have the sensitivity to handle low-ng amount of proteins, along with built-in template methods with recommend parameters to accelerate ease-of-use. Thus, it is realistic to suggest that Orbitrap technology could be deployed not just in core centers, but in general research and clinical research labs for single-cell applications.”
Robitaille adds that MS-SCP and emerging protein sequencing approaches can be complementary technologies. “MS-based proteomics, specifically, is considered the gold-standard for target specificity due to its ability to provide accurate and precise quantitation of proteins,” he explains. “MS offers an unbiased view of the proteome, with the ability to measure various biological functional forms—including peptides, proteins, post-translational modifications, and proteoforms—enabling comprehensive and in-depth analysis. As the field of single-cell omics continues to rapidly evolve, MS uniquely provides the necessary experimental flexibility to adapt and keep up with the pace of change, such as expanding into single-cell metabolomics, lipidomics, and glycomics.”
Bruker

At last year’s ASMS meeting, Bruker noted that its new instrument, the timsTOF Ultra, incorporates a new Captive Spray Ionization (CSI) Ultra ion source with “larger capillary and optimized vortex gas flow, a novel 4th-generation TIMS (trapped ion mobility separation) XR cell and 14-bit digitizer.” The timsTOF Ultra, a company announcement indicated, “can identify over 55K peptides that map into 5,000 protein groups at the single-cell level of 0.125 ng protein loading, at 1% false-discovery rate, and over 4,800 protein groups quantified at CVs of <20%.”
“While single-cell analysis has been game-changing, we face obstacles in maximizing its potential for throughput and protein groups that can be analyzed. It would make biological sense to detect 6,000 or more proteins in a single-cell experiment,” Mechler remarked. “The timsTOF Ultra has overcome these barriers, allowing us to explore the proteome of individual cells at speed and unparalleled sensitivity. Thanks to the timsTOF Ultra, single-cell analysis has reached new heights, and I’m excited to see where this breakthrough will lead us next.”
References
- Sarkar AA. Uncovering Proteomic Patterns One Cell at a Time. GEN 2022; 42(11): 48–51.
- Sarkar AA. Single-Cell Proteomics Bypasses Bottlenecks, Sways Skeptics. GEN 2023; 43(3): 52–55.
- Mechtler K, Hartlmayr D, Matzinger M. New Developments in Mass Spectrometry–Based Single-Cell Proteomics [webcast].
- Hartlmayr D, Ctortecka C, Seth A, et al. An automated workflow for label-free and multiplexed single-cell proteomics sample preparation at unprecedented sensitivity. bioRxiv. April 14, 2021. DOI: 10.1101/2021.04.14.439828.
- Ahmad R, Budnik B. A review of the current state of single-cell proteomics and future perspective. Anal. Bioanal. Chem. 2023; 415(28): 6889–6899. DOI: 10.1007/s00216-023-04759-8.
- Gatto L, Aebersold R, Cox J, et al. Initial recommendations for performing, benchmarking and reporting single-cell proteomics experiments. Nat. Methods 2023; 20: 375–386. DOI: 10.1038/s41592-023-01785-3
- Heumos L, Schaar AC, Lance C, et al. Best practices for single-cell analysis across modalities. Nat. Rev. Gen. 2023; 24: 550–572. DOI: 10.1038/s41576-023-00586-w.
- Orsburn BC. Democratizing Single-Cell Proteomics with Microflow Time-of-Flight Mass Spectrometry [webcast].
- Kaufmann A, Walker S. Comparison of linear intrascan and interscan dynamic ranges of Orbitrap and ion‐mobility time‐of‐flight mass spectrometers. Rapid Commun. Mass Spectrom. 2017; 31: 1915–1926.
- Orsburn BC, Yuan Y, Bumpus NN. Insights into protein post-translational modification landscapes of individual human cells by trapped ion mobility time-of-flight mass spectrometry. Nat. Commun. 2022; 13(1): 7246. DOI: 10.1038/s41467-022-34919-w.


