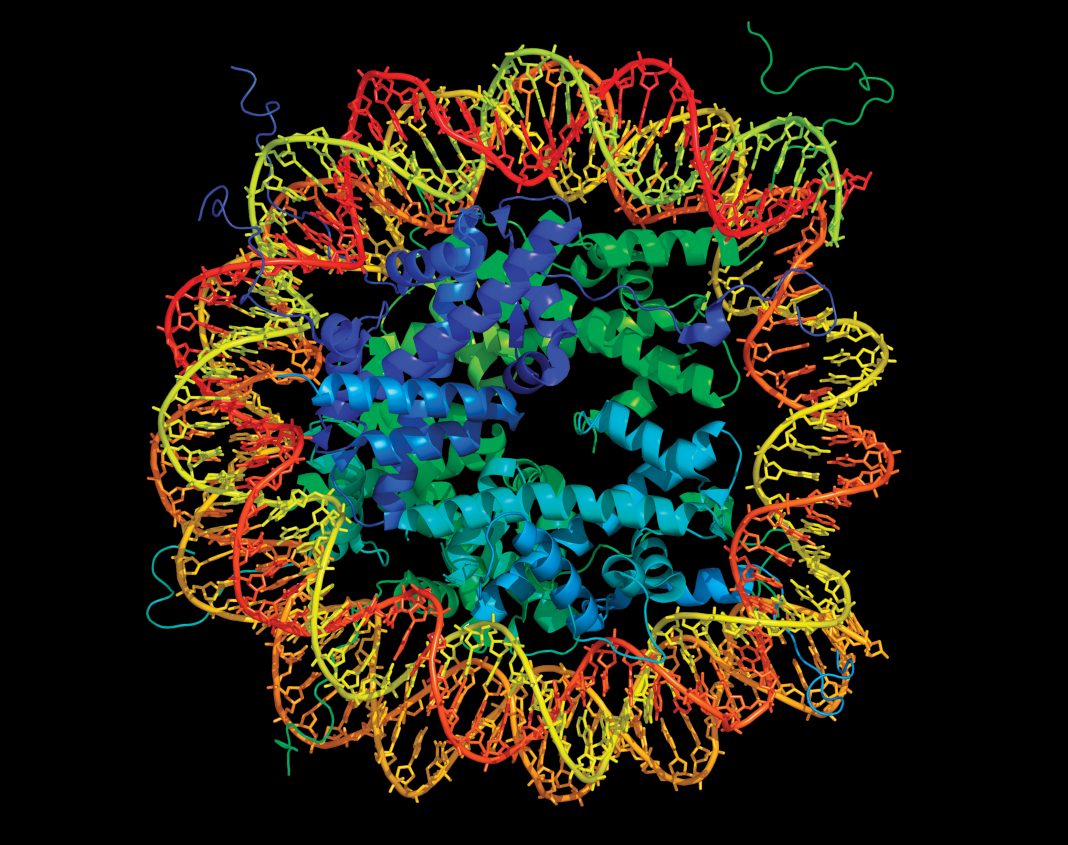
Whether they are partially or fully differentiated, individual cells within multicellular organisms must confront life’s big questions: Who am I? Why am I here? Where am I going? Cells that forget the right answers may succumb to dysfunction and contribute to neurodegenerative, autoimmune, or metabolic disease—or cancer. In the case of cancer, cells may lose themselves in sketchy identities, deviant metabolism, and wanton proliferation, as well as metastatic corruption.
A cell’s fate, be it wholesome or debased, is determined by epigenetics, that is, by patterns of gene expression. Fortunately, normal patterns may be distinguished from aberrant patterns by technologies that interrogate epigenetic factors and monitor epigenetic changes. Even better, if these technologies detect aberrant gene expression, they may prompt pharmacological interventions.
Pharmacological interventions that aim to reverse aberrant gene expression are becoming increasingly specific—and successful. For example, therapeutics are targeting epigenetic regulators of different types, including the so-called writers, readers, and erasers of epigenetic marks. These regulators figure prominently in a growing number of medically relevant cellular pathways.
Epigenetic regulators are subject to modulation by a growing number of epigenetic therapies, some of which have already been approved by the FDA. Other epigenetic therapies are at various stages of preclinical and clinical development.
Selectively inhibiting epigenetic regulators with small molecules
“When a handful of eager young scientists and I founded our company 11 years ago, our core mission was to identify and develop small molecules that inhibit chromatin regulators,” says Patrick Trojer, PhD, chief scientific officer at Constellation Pharmaceuticals. By inhibiting chromatin regulators, small-molecule drugs may, he emphasizes, “manipulate gene expression programs in cancer cells and immune cells.”
Constellation built an integrated epigenetics platform that can validate therapeutic targets and deliver drug candidates that can address unmet clinical needs for malignancies characterized by aberrant gene expression. “After a decade of effort,” Trojer continues, “we are gratified to have two drugs close to achieving clinical proof of concept.” The first targets a bromodomain and extraterminal domain select BET) protein, an epigenetic reader. The second targets the enhancer of zeste 2 polycomb repressive complex 2 subunit select EZH2 union an enzyme that functions as an epigenetic writer. Both drugs are in Phase II clinical trials.
“We are testing our BET inhibitor CPI-0610 in patients with myelofibrosis as a monotherapy or in combination with ruxolitinib,” specifies Trojer. CPI-0610 is being evaluated in MANIFEST, a global study, for its ability to modify, alone or in combination with ruxolitinib, the underlying disease process of myelofibrosis. Positive results would include improvements in hematopoietic function, reductions in spleen volume, and amelioration of patient symptoms. Presentations of interim data may be delivered in the second quarter of the year, and another update may be available in the second half of the year.
The second compound in Phase II clinical trials is CPI-1205, a small-molecule EZH2 inhibitor. “We are exploring CPI-1205 as a second-line drug in a Phase II U.S.-only clinical trial called ProSTAR for patients with metastatic castration-resistant prostate cancer,” informs Trojer. Patients who progressed while on enzalutamide or abiraterone acetate, which block the androgen receptor signaling pathway, are moved to the other of these two first-line drugs and combined with CPI-1205. “We expect to present Phase Ib data at a medical meeting in the second quarter and an interim data update of our Phase II study in the second half of this year,” continues Trojer.
Constellation is also working on a second-generation EZH2 inhibitor, CPI-0209, currently in IND-enabling studies. This molecule is substantially differentiated from first-generation EZH2 inhibitors in its comprehensive target engagement and long duration of action, and a Phase I clinical trial using CPI-0209 for solid tumors is expected to start in mid-2019.
“The biggest challenge in taking full advantage of epigenetics as a target space in oncology is still translational biology,” declares Trojer. This challenge, the translation of basic science findings into the clinic, is common to both epigenetic therapies and traditional oncology drugs. It reflects the impossibility of predicting a priori which patients will respond to a particular drug.
“None of the novel oncology drugs works in unselected patient populations across the community,” Trojer points out. “This has complicated drug development in oncology.”
About two years ago, Constellation made a significant investment to build strong translational capabilities to facilitate the identification of predictive biomarkers and to help select and enrich patient populations in clinical trials. Such strategies are ideally positioned to help identify new mechanisms to enter the market.
One of most promising applications of epigenetic drugs in oncology is in reprograming cancer cells or the tumor microenvironment to make them more susceptible to other existing drugs or those under development. “While there is still a lot of work to be done,” Trojer states, “Constellation is well positioned to lead the effort of learning about the key epigenetic mechanisms in several cancer contexts and applying that learning to bring innovative medicines to patients faster.”
Modulating the epigenetic determinants of synaptic plasticity
“We are particularly interested in synaptic integrity in the brain, which is a primary opportunity and challenge for the field of neuroscience,” says Adam J. Rosenberg, president and CEO of Rodin Therapeutics. “As part of this work, we continue to seek the validation of translational markers.”
Synaptic loss of function or dysfunction drive symptoms in many neurodegenerative, neuropsychiatric, and neurodevelopmental conditions. To preserve or restore synaptic function—that is, to strengthen existing synapses and promote the creation of new synapses—Rodin focuses its drug discovery efforts on modulating neuronal gene expression.
Multiple biomarkers can be used to validate target engagement and functional efficacy. “We now have tools that were not available to us even two or three years ago,” notes Rosenberg.
Rodin is positioning its drug development program to benefit patients with a range of conditions related to synaptic function. “As part of our disease-agnostic approach, we are initially focusing on neurodegeneration,” Rosenberg explains. “[We] hope to later enter other areas, including psychiatric conditions.”
[National Human Genome Research Institute]
The molecular strategy at Rodin relies on the selective inhibition of specific histone deacetylases select HDACs union which constitute a key class of proteins that regulate synaptogenesis and synaptic plasticity, to increase gene expression with minimal safety concerns. Eighteen human HDACs have been described, and while they are attractive drug targets, their therapeutic modulation has been historically complicated by the potential of adverse effects.
“Designing molecules that are complex-selective allows us to disassociate the main class toxicity concern, which has always been the rate-limiting factor for progressing HDAC inhibitors in the clinic,” states Rosenberg. At Rodin, molecules have been identified that specifically and selectively target histone deacetylases as part of the coREST co-repressor complex, which includes HDAC1 and HDAC2.
Rodin’s lead compound, RDN-929, shows a favorable preclinical safety profile and pro-synaptic pharmacological effects at the molecular, structural, and functional levels. RDN-929 increased the levels of synaptic density markers and the number of dendritic spines in a dose-dependent manner in multiple species.
In mice models of neurodegenerative diseases, RDN-929 improved synaptic function, indicating benefits for learning and memory. RDN-929 is now being evaluated in a Phase I clinical trial. “We are hoping that the Phase I safety and translational data will continue to allow us to make smarter drug discovery and development decisions,” says Rosenberg.
Closing off aberrant pathways of proliferation
Heading the Cancer Cell Cycle Group at the University of Oxford, professor of cancer biology Nicholas La Thangue, PhD, studies the molecular pathways that control cell proliferation. His group is particularly interested in how these pathways become aberrant in cancer.
For example, La Thangue’s group has been scrutinizing the pathways that govern the G1-to-S transition. One of these pathways is controlled by the retinoblastoma tumor suppressor protein pRb. Another one is controlled by the p53 tumor suppressor protein. Mutated, dysfunctional versions of pRb and p53 occur in many tumors. Loss of proper pRb function deregulates proliferation. Loss of proper p53 function causes an insensitivity to checkpoint control.
“We believe that biological knowledge on the mechanisms which drive cancer cell proliferation can be harnessed in designing new therapeutic modalities to treat cancer,” states the Cancer Cell Cycle Group’s webpage. “Consequently, we work closely with the biotechnology and pharmaceutical sectors…in translating our academic discoveries into an applied clinical setting.” Directly bridging the academic/commercial divide, La Thangue serves as the CEO of Celleron Therapeutics and the CSO of Argonaut Therapeutics.
“Celleron’s clinical trials are focusing on an epigenetic regulator that was initially identified as a histone deacetylase inhibitor,” says La Thangue. “In preclinical studies, we later uncovered that it also functions as an immune regulator.”
CXD101, Celleron’s proprietary asset and one of the two compounds in Phase II trials, is a dual-mechanism HDAC inhibitor that increases the immune recognition and destruction of malignant cells. In preclinical studies, CXD101 showed promise as a monotherapy in lung and colon xenografts, and in Phase I trials, it led to tumor remission. CXD101 is said to have a dual mechanism of action because the drug has two abilities: it can reestablish normal function in tumor cells, and it can reinstate immune relevant gene expression.
As part of epigenetic drug discovery efforts at Argonaut, La Thangue and colleagues are developing inhibitors of protein arginine methyltransferase to target one of the newest levels of epigenetic control, arginine methylation. This group of cellular enzymes is involved in gene transcription, RNA splicing, DNA repair, and protein translation. These enzymes have also been linked to malignant transformation and metastasis.
“Arginine methylation was more recently described than acetylation,” notes La Thangue. “It represents a novel and exciting level of epigenetic control, and small molecules that are being developed to target this epigenetic modification have shown quite profound effects in the clinic.”
Scientists at Argonaut reported that arginine methylation is an important mechanism involved in cancer cell proliferation, and efforts are underway to develop inhibitors of arginine methylation that reinstate cellular suicide pathways and stop cancer cell growth.
“The real challenge with any cancer drug, but particularly with epigenetic regulators, is that the pathways that are being targeted are usually deregulated in only a small subset of the tumors,” observes La Thangue. Efforts to therapeutically target specific subsets of cancer patients with individual drugs depend on the identification of biomarkers that could predict the response to therapy.
“At Celleron, we are using precision medicine approaches and genome-wide technologies to identify positive predictors of clinical response,” states La Thangue. Doing so, he continues, may help the company “target the right patients with the right tumors at the right time.”
In addition, there is a need to understand the benefits and limitations of using drug combinations. “Understanding the defects in epigenetic pathways would provide an opportunity to combine the right drugs,” emphasizes La Thangue. “That is a challenge led by genome-wide studies together with transcriptomic and proteomic approaches to help use that data in the most sensible way to identify the right combination.”
Capturing the epigenetic dimension of cancer cell heterogeneity
To capture cell-to-cell heterogeneity in populations of cells, scientists often make use of single-cell RNA sequencing select RNA-seq). Although single-cell RNA-seq is sensitive to heterogeneity at the transcriptional level, it may miss subtleties of heterogeneity at the epigenetic level. Consequently, scientists have developed techniques that are more epigenetically focused.
For example, scientists based at the University of Stamford developed a method for integrative epigenomic analysis called the Assay for Transposase Accessible Chromatin with high-throughput sequencing select ATAC-seq). The method, which relies on in vitro transposition of sequencing tags into areas of accessible chromatin, can allow simultaneous, genome-wide information on the positions of open chromatin, transcription factor binding, and nucleosomes in regulatory regions, as well as information on chromatin state annotation.
To develop ATAC-seq commercially, several Stamford University researchers select Paul Giresi, PhD, Howard Chang, PhD, and William Greenleaf, PhD) helped found Epinomics. The company proceeded to demonstrate that ATAC-seq could be used to advance personalized medicine.
In late 2018, Epinomics was acquired by 10x Genomics. Subsequently, Epinomics’ ATAC-seq technology was incorporated into 10x Genomics’ first epigenomics-focused product, the Chromium Single Cell ATAC Solution. Now the product is being adopted across research areas such as oncology, immunology, and drug development.
The Chromium Single Cell ATAC Solution can profile hundreds to thousands of cells in a single reaction and is scalable for high-throughput studies. “This allows us to profile open chromatin regions in cells and capture promoter and regulatory region information in a single assay,” says Laura DeMare, PhD, associate product manager of epigenomics at 10x Genomics.
The solution, which includes reagents, a microfluidic chip, and analysis software, allows investigators to generate sequencing libraries of 500–10,000 cells from a biological sample and run the sequencing data through the bioinformatics pipeline to get a cell-by-cell readout of accessible chromatin sites. “We can identify the transcription factor motifs at accessible chromatin sites,” asserts DeMare. “That gives a sense as to the potential master regulators that are influencing gene expression in that cell.”
The solution’s single-cell resolution analyses are particularly informative for cancer biology, given the cell-to-cell heterogeneity that characterizes many cancers. “Looking at the single-cell level is critical,” insists DeMare. “We know that there is heterogeneity in chromatin accessibility within tumor samples.”
ATAC-seq is also relevant to therapeutic development. For example, ATAC-seq can be applied to interrogate the mechanisms that cancers exploit to develop drug resistance. “Many cancer drugs are not always fully effective, and that happens because of the heterogeneities among the cancer cells,” notes DeMare. Single-cell chromatin accessibility analyses, by capturing single-cell chromatin states, can shed light on how and why cells representing a small subset of cells within a tumor become drug resistant. According to DeMare, “ATAC-seq can provide an additional dimension to disentangle mechanisms of acquired resistance, which RNA-seq may not reveal.”
Developing RNA-Level Epigenetic Therapeutics
DNA and histones are not the only structures that may gain or lose epigenetic marks. RNA, too, has its modifications, and they provide an additional level of epigenetic control. At this level, the epitranscriptomic level, RNA modifications may contribute to health or disease. Consequently, drug developers are harnessing epitranscriptomics, or RNA epigenetics, to modulate biological processes and thereby effect new treatments.
For example, Storm Therapeutics is investigating how RNA epigenetic drugs could be used to treat cancer and other diseases. To do so, the company is taking advantage of recent technical advances in mass spectrometry and RNA sequencing. These technologies are helping Storm interrogate RNA modifications and characterize their contribution to biological processes. Storm’s efforts, suggests Keith Blundy, PhD, the company’s director and CEO, are facilitated by an increasing understanding of the biological functions of RNA and their implications in disease.
“We are trying to inhibit the enzymes that make modifications on RNA, such as methylation,” says Blundy. With this comment, Blundy makes it clear that Storm’s efforts are not directed toward developing RNA-binding drugs. Instead, the company is targeting enzymes that may act upon RNA molecules, altering their behaviors select which may, incidentally, include protein binding).
“When we started, we did not know whether RNA methyltransferases would be chemically tractable,” recalls Blundy. “But now we know that it is possible to get very potent and selective inhibitors of RNA methyltransferases.”
Like epigenetic modifications on DNA and histones, RNA epigenetic modifications are orchestrated by the same broad types of molecules, including writers, erasers, and readers of epigenetic information. “Much less is known about all these complexes than in the case of DNA epigenetic changes,” notes Blundy. Storm is focusing on writers of RNA epigenetic marks. “We think this is the most druggable space,” says Blundy.
The existence of multiple species of RNA, and of many more types of chemical modifications than for DNA and histones, opens opportunities to modulate gene expression using additional and subtler layers of epigenetic regulation. “Another reason why RNA epigenetic modifications might be an attractive intervention point is that, as compared to DNA and histone epigenetic modifications, they are less likely to change the gross structure of the chromosome,” explains Blundy. Changes in gross structure could result in off-target effects.
A challenge in the development of therapeutics that target RNA methyltransferases is the need to develop and perform biochemical assays. Such work can be time-consuming. Another challenge is the need to measure individual modifications on the target RNA in a comprehensive manner, not only in cells but also in tissue samples. “There is no universal way of doing that,” observes Blundy.
Different studies have used antibodies, pull-down methodologies, or RNA-seq to capture these modifications. “The only tool that gets close to a universal approach is mass spectrometry,” states Blundy. “We have invested significantly into our platform to measure these modifications in a sequence-specific context with enough sensitivity.”


