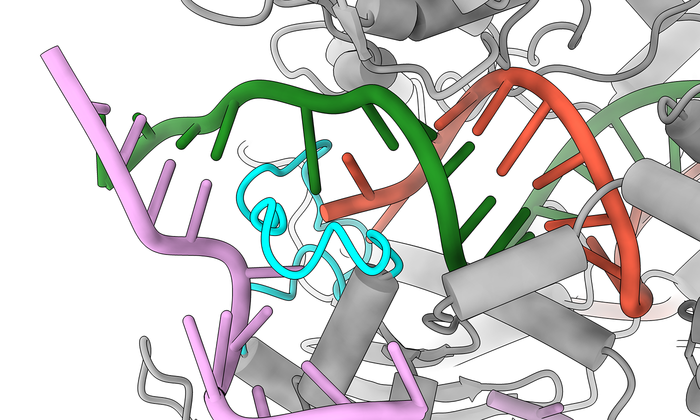
CRISPR–Cas9 has limitations. One of the most concerning, in regards to using genome editing for therapeutic applications for humans, is off-target DNA cleavage. Although some Cas9 variants have been developed that have improved mismatch discrimination, they suffer from reduced rates of on-target DNA cleavage.
Because the underlying mechanisms by which Cas9 recognizes mismatches are poorly understood, a group of scientists sought to determine the structure of Cas9 at different stages of mismatch cleavage using kinetics-guided cryo-electron microscopy. In doing so, they provided a molecular blueprint for the design of next-generation high-fidelity Cas9 variants that reduce off-target DNA cleavage while retaining efficient cleavage of on-target DNA. Using that blueprint, they designed a high-fidelity variant that retains wild-type on-target cleavage rates which they call “SuperFi-Cas9.”
The work is described in a paper published in Nature titled, “Structural basis for mismatch surveillance by CRISPR–Cas9.”
“This really could be a game-changer in terms of a wider application of the CRISPR Cas systems in gene editing,” said Kenneth Johnson, a professor of molecular biosciences at the University of Texas (UT), Austin.
SuperFi-Cas9 is 4,000 times less likely to cut off-target sites but just as fast as naturally occurring Cas9. Jack Bravo, PhD, a postdoctoral fellow in the lab of David Taylor, PhD, assistant professor in the department of molecular biosciences at UT Austin, said you can think of the different lab-generated versions of Cas9 as different models of self-driving cars. Most models are really safe, but they have a top speed of 10 miles per hour. “They’re safer than the naturally occurring Cas9, but it comes at a big cost: They’re going extremely slowly,” said Bravo. “SuperFi-Cas9 is like a self-driving car that has been engineered to be extremely safe, but it can still go at full speed.”
So far, the researchers have demonstrated the use of SuperFi-Cas9 on DNA in test tubes. They’re now collaborating with other researchers who plan to test SuperFi-Cas9 for gene editing in living cells. They’re also working to develop still safer and more active versions of Cas9.
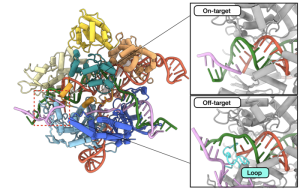
Taylor and Johnson developed a technique called kinetics-guided structure determination that used a cryo-electron microscope in the Sauer Structural Biology Lab to image Cas9 in action as it interacted with mismatched DNA.
“It’s like if you had a chair and one of the legs was snapped off and you just duct taped it together again,” Bravo said. “It could still function as a chair, but it might be a bit wobbly. It’s a pretty dirty fix.”
Without that added stability in the DNA, Cas9 doesn’t take the other steps needed to cut the DNA and make edits. No one had ever observed this extra finger doing this stabilization before.
“This was something that I could never have, in a million years, imagined in my mind would have happened,” Taylor said.
Based on this insight, they redesigned the extra finger on Cas9 so that instead of stabilizing the part of the DNA containing the mismatch, the finger is instead pushed away from the DNA, which prevents Cas9 from continuing the process of cutting and editing the DNA. The result is SuperFi-Cas9, a protein that cuts the right target just as readily as naturally occurring Cas9 but is much less likely to cut the wrong target.