Ideally, fluorescently labeled cell signaling events that occur all at once, in a flurry, should also be recorded all at once. But there’s a problem. Different kinds of cell signals are usually distinguished with spectral multiplexing, which depends on spectrally distinct reporter molecules, such as green fluorescent protein, which aren’t, alas, as spectrally distinct as we might wish. So, to assist spectral multiplexing, MIT scientists developed a spatial multiplexing technique. In combination, spectral multiplexing and spatial multiplexing allow multiple reporters to be imaged—and distinguished—at once.
Actually, it would be more accurate to say that with the new technique, a spectrally multiplexed image may be aligned with a spatially multiplexed image. The former is captured first with live-cell imaging; the latter is captured next, with a fixed-cell imaging technique such as serial antibody staining. The combination works because, as the MIT scientists have demonstrated, the labeled entities remain stationary for at least an hour, long enough for spatial multiplexing to be brought to bear.
Details of the combination technique appeared November 23 in the journal Cell, in an article titled, “Spatial Multiplexing of Fluorescent Reporters for Imaging Signaling Network Dynamics.” This article described how different fluorescent reporters may be fused to different pairs of self-assembling peptides to signal reporter islands (SiRIs). Stochastically clustered at different points in space, separated by micrometers, the labeled entities known as SiRIs enable dense sampling of signals throughout cells.
According to the study’s authors, SiRIs may be “distant enough to be resolved by a microscope, but close enough to spatially sample the relevant biology.”
“Because [SiRIs] can be modularly designed, they permit a set of fluorescent reporters to be efficiently adapted for simultaneous measurement of multiple nodes of a signal transduction network within single cells,” the authors continued. “We created SiRIs for indicators of second messengers and kinases and used them, in hippocampal neurons in culture and intact brain slices, to discover relationships between the speed of calcium signaling, and the amplitude of PKA signaling, upon receiving a cAMP-driving stimulus.”
In the current study, which was led by MIT’s Edward Boyden, PhD, up to five different molecule types were imaged simultaneously by measuring each signal from random, distinct locations throughout a cell. This approach could allow scientists to learn much more about the complex signaling networks that control most cell functions.
“There are thousands of molecules encoded by the genome, and they’re interacting in ways that we don’t understand,” said Boyden. “Only by watching them at the same time can we understand their relationships.”
Boyden and his colleagues demonstrated their technique by using it to identify two populations of neurons that respond to calcium signals in different ways. These signal responses, the scientists indicated, may influence how long-term memories are encoded.
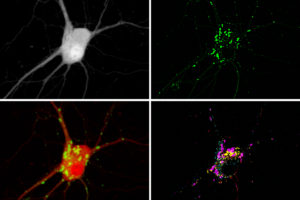
To make molecular activity visible within a cell, scientists typically create reporters by fusing a protein that senses a target molecule to a protein that glows. “This is similar to how a smoke detector will sense smoke and then flash a light,” noted Shannon Johnson, an MIT graduate student and one of the study’s lead authors.
“Typically a biologist can see one or two colors at the same time on a microscope, and many of the reporters out there are green, because they’re based on the green fluorescent protein,” Boyden added. “What has been lacking until now is the ability to see more than a couple of these signals at once.”
“Just like listening to the sound of a single instrument from an orchestra is far from enough to fully appreciate a symphony,” remarked MIT postdoc Changyang Linghu, PhD, the study’s other lead author. “By enabling observations of multiple cellular signals at the same time, our technology will help us understand the ‘symphony’ of cellular activities.”
To boost the number of signals they could see, the researchers set out to identify signals by location instead of by color. They modified existing reporters to cause them to accumulate in clusters at different locations within a cell. They did this by adding two small peptides to each reporter, which helped the reporters form distinct clusters within cells.
“It’s like having reporter X be tethered to a LEGO brick, and reporter Z tethered to a K’NEX piece—only LEGO bricks will snap to other LEGO bricks, causing only reporter X to be clustered with more of reporter X,” Johnson explained.
With this technique, each cell ends up with hundreds of clusters of fluorescent reporters. After measuring the activity of each cluster under a microscope, based on the changing fluorescence, the researchers can identify which molecule was being measured in each cluster by preserving the cell and staining for peptide tags that are unique to each reporter. The peptide tags are invisible in the live cell, but they can be stained and seen after the live imaging is done. This allows the researchers to distinguish signals for different molecules even though they may all be fluorescing the same color in the live cell.
To demonstrate the potential usefulness of their strategy, the scientists measured the activities of three molecules in parallel—calcium, cyclic AMP, and protein kinase A (PKA). These molecules form a signaling network that is involved with many different cellular functions throughout the body. In neurons, it plays an important role in translating a short-term input (from upstream neurons) into long-term changes such as strengthening the connections between neurons—a process that is necessary for learning and forming new memories.
The researchers now plan to try their approach in living animals to study how signaling network activities relate to behavior. The researchers also intend to expand their signal-deciphering work to other types of cells, such as immune cells. The new technique, the researchers suggest, could also be useful for comparing signaling network patterns between cells from healthy and diseased tissue.
In this paper, the researchers asserted that by modifying their existing strategy, they could record up to 16 different molecular signals at once. With additional work, that number could reach into the hundreds, the researchers believe.
“That really might help crack open some of these tough questions about how the parts of a cell work together,” Boyden speculated. “One might imagine an era when we can watch everything going on in a living cell, or at least the part involved with learning, or with disease, or with the treatment of a disease.”


