Sponsored content brought to you by

The importance of genetic stability in mouse research
To the average life-science researcher, a mouse’s genetic background may be an afterthought, if even a thought, at all. A researcher’s top priorities may be to understand disease, to publish, and to obtain funding. However, to successfully reach these goals, maintaining genetic stability, or preventing genetic drift, in a mouse colony should be of high importance.
Laboratory mice are unique, live elements in scientific research that change over the course of their lifetime, and importantly, from one generation to the next. After all, heritable changes in DNA sequence are the basis for species diversity and evolution in the wild. Even in the absence of evolutionary pressure, changes in DNA sequence occur. At first glance, these mutations seem to be silent, unimportant fluctuations in the genetic makeup of an individual. However, these seemingly insignificant mutations can become the source of unexplainable experimental irreproducibility.
Mouse researchers, then, are met with a conundrum. Generating mice for research requires breeding. But, with breeding comes the inherent risk to propagate genetic diversity and thus, to propagate experimental diversity. From one experiment to the next and from one publication to the next, data diversity is unconducive to scientific progress.
The purpose of this paper is to educate mouse researchers on the potential for genetic drift to impact research progress, to highlight best practices to minimize drift, and provide solutions to reverse drift if it arises in a mouse colony. Use of full official mouse strain nomenclature and careful reporting of breeding generation information in publications and grant proposals are some simple practices researchers can take that promote reproducibility and responsible animal use.
How genetic drift arises and its prevalence in mouse colonies
Inbreeding, or sibling mating, is a powerful method to reduce heterozygosity at every genetic locus in the mouse genome, allowing for uniformity in phenotype and forming the basis for experimental reproducibility. Genetic homozygosity allows comparison of a single variable between a control and an experimental group, and thus, to be able to attribute any differences in readout to that variable.
Much like species in the wild, two populations of inbred laboratory mouse strains maintained in isolation from each other will change over time. Spontaneous mutations may occur in the form of single nucleotide polymorphisms (SNPs), deletions, inversions, duplications, and other such errors during DNA replication and meiosis. This process of spontaneous mutations appearing, disappearing, or becoming fixed in a population at random is called genetic drift (Lee Silver, 1995).
The amount of genetic drift occurring in any actively breeding colony varies, but is predicted to be rather frequent. The average breeding generation is 3-4 months long with mice becoming sexually mature around 5-8 weeks of age. Offspring are typically born about 3 weeks after mating. Based on spontaneous mutation rates calculated from coat color mutations measured in over 1 million mice, 1 phenotypic mutation may arise every 1.8 breeding generations (Drake et al., 1998; Russell and Russell, 1996).
The risk of breeding a mouse that carries a spontaneous mutation in the germ line, and thus of propagating this mutation, is higher in small colonies than in large colonies (Figure 1A). For any given germ line mutation in a mouse, roughly half of its offspring will be heterozygous for this mutation (Figure 1B). In inbred breeding colonies, there is a 25% chance these mutations will become fixed (homozygous) in the population (Chamary and Hurst, 2004; Drake et al., 1998).
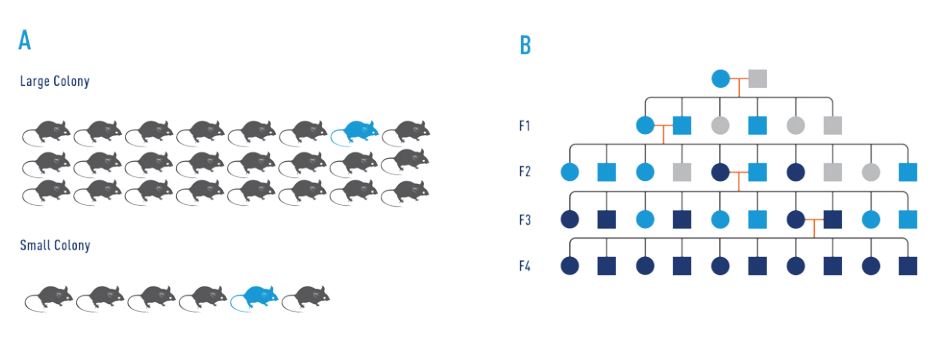
Indications that genetic drift has occurred: Substrain designations

A substrain is a branch of an inbred strain that is suspected or known to be genetically different from the parent colony (www.informatics.jax.org/mgihome/nomen/strains.shtml#substrains). Because genetic drift may differentially occur in two populations of any given inbred strain, substrain designation is a crucial component of nomenclature. Substrains are designated by adding a unique lab code assigned by the Institute for Laboratory Animal Research (ILAR) (dels.nas.edu/global/ilar/Lab-Codes). A lab code identifies the institute, lab, or investigator that produced or maintains a particular animal strain (Table 1). Because lab codes accumulate in the nomenclature, the strain’s genealogy is understood from the name alone. For example, strain C57BL/6NJ was maintained for many years at the National Institutes of Health (N) and is now distributed by The Jackson Laboratory (J) (Figure 3). By extension, the substrain nomenclature gives a general indication that genetic variation between two strains exists.
Suspected genetic differences: Generation number
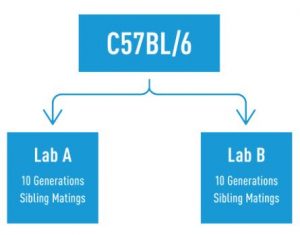
Any strain that has been maintained separately from the parental strain for 20 consecutive inbred generations (~5-6 years) is suspected to carry genetic differences, and is therefore considered a substrain. Additionally, breeding generations are cumulative, such that if two labs obtain mice from the same common ancestor and breed for 10 generations, each lab has a different substrain from one another because the two strains are considered 20 generations apart (Figure 2).
The very first inbred mouse strains (including C57BL/6, DBA, C3H, BALB, CBA, and others) used for biological research were established almost 100 years ago and continue to be heavily published today. Because these strains exceed 200 inbred generations and because multiple institutions worldwide breed them, a considerable amount of genetic drift has occurred over time in all of these strains. Because of genetic drift, it is possible that observations made in existing substrains differ from observations made in the parental inbred strains from which they were derived.
Known genetic differences: Substrain designation by observed phenotypic differences
Additionally, substrains are designated when a difference in phenotype is observed between two groups of inbred mice. However, unless these spontaneous mutations manifest obvious phenotypes, frequently after they become fixed homozygous in the colony, and attentive colony managers or researchers recognize something “off” about the mice, the mutations may be carried in a strain unnoticed for years. Thus, identifying drift may depend on individual labs asking questions whose answers happen to rely on such mutations, to recognize that “unexpected results” are more than just “failed experiments,” and later to identify the mutation that is responsible for the aberrant phenotype.
For example, the parental inbred strain C3H gave rise to two substrains from two Jackson Laboratory researchers, which for many years, did not seem to differ. Dr. Walter Heston bred the strain in the 1930’s (now C3H/HeJ). In 1952, Heston transferred some of his mice to another Jackson Laboratory researcher, Dr. Henry Outzen (now C3H/HeOuJ). In the late 1960s, Heston’s strain was found to be resistant to lipopolysaccharide (LPS), whereas Outzen’s strain remained sensitive.
Later, the mutation was mapped to Tlr4, a gene involved in pathogen recognition and innate immune system activation (Poltorak et al., 1998a; Watson et al., 1978). By the time the C to A substitution at nucleotide 2342 was identified in Tlr4, it had already become fixed in the Heston substrain, likely between 1958 and 1965 (Poltorak et al., 1998b). Had Heston’s C3H substrain never been treated with LPS, it is possible that the Tlr4 mutation would not have been identified and conclusions involving basic immunology in these strains may have become highly controversial.
Known genomic sequences are substrain-specific
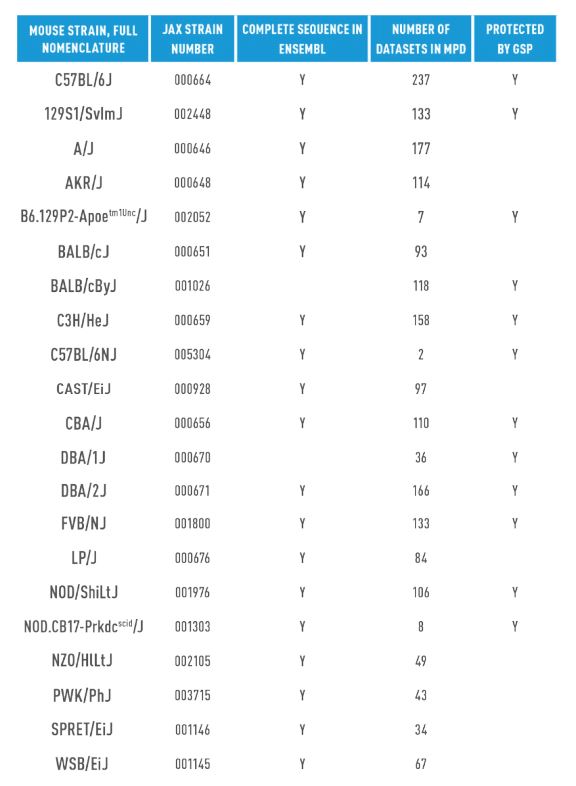
Aside from chance discoveries, the only way to definitively identify whether genetic drift has occurred is to sequence the strain and compare to reference genomes. A C57BL/6J female was the first mouse to be completely sequenced by the Mouse Genome Sequencing Consortium (Chinwalla et al., 2002), www.ensembl.org/Mus_musculus). To date, 15 other major inbred mouse strains have been fully sequenced, all of which are “J” substrains, the official ILAR laboratory code for The Jackson Laboratory (Adams et al., 2015), www.ensembl.org/Mus_musculus/Info/Strains) (Table 2).
An additional 20+ inbred strains have been sequenced using short-read approaches to identify SNPs, indels, and structural variations relative to the C57BL/6J mouse reference genome (Frazer et al., 2007 and www.sanger. ac.uk/science/data/mouse-genomes-project). Furthermore, known SNP data for specific substrains can be found and compared in the Mouse Phenome Database (MPD), a collaborative standardized collection of genotypic and phenotypic data on the most commonly published mouse strains (phenome.jax.org).
Genetic background impacts research conclusions
As described earlier with the C3H example, substrains may acquire spontaneous mutations that have the potential to influence research conclusions. If these experiments are not properly controlled for, such as through use of appropriate substrain, disastrous consequences on experimental reproducibility may ensue. Whether these spontaneous mutations arise in a repository, from a vendor, or in individual laboratories, how can researchers know which is the “best” substrain to use for their experiments?
Unfortunately, there isn’t an easy answer. The best way to determine whether genetic background matters is to perform controlled, side-by-side experiments and compare. Since it is impossible to test every substrain that exists for a particular experimental readout, the next best way to understand the potential impact of genetic background on research conclusions is to rely on what other researchers have observed, in the form of peer-reviewed, published literature and to continue experiments that build on such knowledge using identical substrains.
Conclusion
Genetic drift is an inevitable reality in actively breeding mouse colonies and may deeply impact research conclusions and reproducibility. While genetic drift cannot be eliminated completely, colony management strategies can be implemented both in individual laboratories and in large mouse repositories and vendors to maintain genetic stability. Reproducibility and scientific discovery rely on careful reporting of complete mouse substrain 17


