The protein hub known as WD40 repeat protein 5 (WDR5) participates in complex, multistep activities such as epigenetic regulation and cell division. To put it bluntly, WDR5 is a multitasker. However, like many multitaskers, it operates in ways that are hard to follow. So, when it malfunctions, its slip ups can be as hard to analyze as the mistakes of an almost perfect juggler.
Whatever the juggled objects happen to be, the slip ups or mistakes come down to poor timing. Consider the slip ups that may occur with WDR5—slip ups that may give rise to disease states such as cancer. In these cases, too little or too much time may elapse during association or dissociation between proteins.
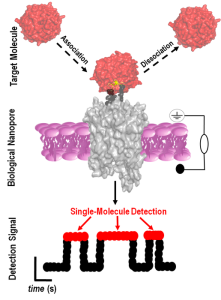
To better understand WDR5’s transient interactions with other proteins, researchers at Syracuse University engineered a highly sensitive nanopore sensor. The researchers assert that their nanopore sensor can detect WDR5 with single-molecule fidelity in solution. They add that they have used their nanopore sensor, which is named MLL4WintFhuA, to study multimodal protein recognition events.
The researchers detailed their findings in Nature Communications, in an article titled, “Disentangling the recognition complexity of a protein hub using a nanopore.” They described how their nanopore sensor incorporated a monomeric β-barrel scaffold of ferric hydroxamate uptake component A (FhuA) from Escherichia coli (a construct named tFhuA) and a 14-residue Win motif of mixed lineage leukemia 4 (MLL4Win) methyltransferase, a WDR5 ligand.
“Our approach reveals a broad dynamic range of MLL4Win-WDR5 interactions and three distant subpopulations of binding events, representing three modes of protein recognition,” the article’s authors wrote. “The three binding events are confirmed as specific interactions using a weakly binding WDR5 derivative and various environmental contexts.”
This work was funded through a four-year, $1.2 million Research Project Grant (R01) from the National Institutes of Health’s National Institute of General Medical Sciences (NIGMS), awarded to Liviu Movileanu, PhD, professor of physics, in 2018. The research team also includes Lauren Ashley Mayse and Ali Imran, both graduate students in Movileanu’s laboratory, as well as other researchers at SUNY Upstate Medical University, Ichor Therapeutics, and the National Institutes of Health’s National Institute of Child Health and Human Development.
The team’s work culminated in the creation of an ultrasensitive device capable of detecting and quantifying WDR5. The researchers designed, developed, and validated a biosensor that consists of a tiny hole (nanopore) in a synthetic membrane and that is capable of identifying proteins in solution at single-molecule precision.
The biosensor’s channel-like base allows ionic solution to flow through it. A change in the ionic flow serves as a signal for the presence of a specific molecule, in this case, WDR5.
“The idea behind was to design nanopores that are equipped with hooks that pull certain proteins from a solution,” said Movileanu, who is also a member of the BioInspired Institute. “By being able to fish them from a solution one at a time, we can better understand how these proteins function.”
The team revealed new details about the conditions under which WDR5 associates with and dissociates from other proteins. Such details will allow researchers to better understand how WDR5 and other multitasking molecules carry out their various responsibilities.
“Proteins need to talk to each other for brief periods,” Movileanu explained. “In the majority of cancers, you have a situation where at least one protein sits on another protein or talks to another protein for much longer than needed. Many biotechnology companies want to develop drugs that perturb those interactions.”
Mayse shares that their study uncovered new information about WDR5’s unique interface, where a peptide must wiggle into its deep and donut-shaped cavity. “We found that our sensor can recognize WDR5 with a weak connection, a medial connection, and a strong connection to a peptide,” she pointed out. “This shows that a potential drug must be able to prevent all three different ways a peptide can associate with WDR5.”
Biosensors like the one developed in Movileanu’s laboratory could one day lead to more accurate and efficient methods of scanning chemicals in the body, providing an opportunity for doctors to detect diseases much earlier than is possible today.
“In many diseases there are markers, chemicals in our body that change quite a bit in a noticeable way when diseases, such as cancer, start to develop,” Movileanu stressed. “By integrating these sensors into nanofluidic devices that are scalable, we are not too far from being able to scan many markers from a sample of blood.”


