Sponsored content brought to you by
The advent of sophisticated cellular imaging techniques, combined with technological advances in automation and data collection and interpretation, has provided the basis for the development of High-Content Screening (HCS), which can give information on how complex cellular functions respond to treatment with pharmacological agents in a high-throughput manner. Coupled with the development of increasingly complex and disease-relevant cellular models, HCS can be applied to support several different steps in the drug discovery continuum, including but not limited to screening, hit identification, hit-to-lead campaigns and structure-activity relationship studies, as well as assessment of ADME and toxicology properties of molecules in relevant cellular models.
IRBM’s expert drug discovery team has significant experience in implementing HCS, developing disease-relevant cellular models and combining the two approaches to support integrated drug discovery research plans with the goal of selecting the best preclinical and/or clinical candidates.
Screening and investigating leads using HCS and automated image analysis
Alberto Bresciani, Head of High Throughput Biology and Screening (HTBS) at IRBM, leads an experienced team dedicated to the intersection of biology and advanced screening technologies to help identify and validate hits and progress them to lead identification and optimization. The HTBS team develops assays quickly and at scale, while also ensuring reproducibility and consistency of experimental setups and results.

“From confocal microscopy to live cell imaging, we offer a wide array of high-content approaches and instruments that can help partners answer almost any question they might have in drug discovery, through multiparametric high-content assays,” states Bresciani. “From target to hit identification, lead optimization and pre-clinical testing, HCS can play a significant role through a variety of imaging and analysis capabilities including the analysis of 2D or 3D cell culture models. The possibilities are almost endless,” comments Bresciani.
‘Cell painting’ is a morphological profiling assay which can be used to analyze various cellular compartments in a single experiment. This technique is a high-content, multiplexed image-based assay where up to six fluorescent dyes are used to label (‘paint’) cell components such as the nucleus, endoplasmic reticulum, mitochondria, cytoskeleton and Golgi apparatus, as well as RNA. By applying automated image analysis, features can be measured and compared within each cell studied.
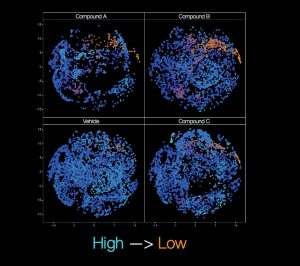
Bresciani shares one example of how cell painting has been used in Duchenne Muscular Dystrophy (DMD) research. “We tested approximately 10,000 cell-active compounds to look for any impact on adipocytic versus fibrotic cell commitment.” Imaging plus determination of single-cell features can be calculated to provide highly insightful results. “We use image pre-processing with basic segmentation and target linking, followed by calculation mapping and further processing to determine sub-populations and from that, identify the active compounds,” describes Bresciani.
“In our DMD studies, we worked with our collaborators to show that the muscle micro-environment is able to influence muscle fibro/adipogenic progenitor cell response to gluco-corticoids via cell autonomous and non-autonomous signalling, as well as how these progenitors can also be affected by the WNT5a/GSK3/β-catenin axis.” The results of the researchers’ studies were published in Scientific Reports(1) and Cell Death and Differentiation(2). Image analysis, which is key to approaches such as cell painting, therefore provides a versatile tool in providing reliable and statistically robust quantitative data.
“When it comes to image analysis, IRBM employs automated and advanced high-content image analysis software that provides advanced segmentation, pre-processing and post-processing tools,” adds Bresciani.
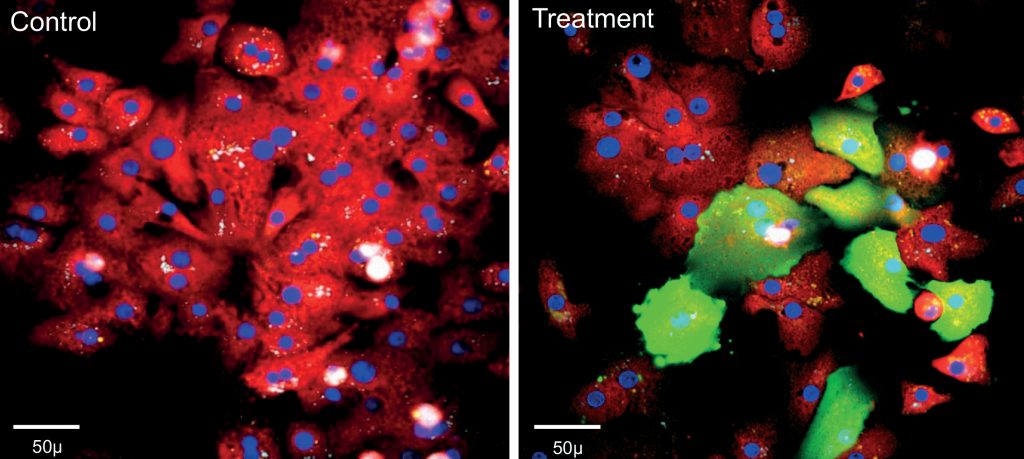
Establishing pathophysiologically relevant cellular models
To leverage the full potential of HCS, it is important to apply the platform to relevant cellular models. Dr. Annalise Di Marco, Head of ADME/Pharmacology at IRBM, describes her team’s experience with the establishment of assays capable of predicting the potential liver toxicity of drug candidates. “Hepatoxicity is still one of the major causes for drug failure in the late stage of development when not in the clinic. Primary 2D human hepatocyte cultures have a limited predictive value because they dedifferentiate very rapidly within hours after plating and lose important hepatic functions: therefore, only short-term treatment studies can be performed with the molecule under investigation. However, more accurate assessment can be made by applying an HCS-based approach to 3D cultures, such as liver spheroids, treated for multiple days with the drugs of interest and assessing various effects such as apoptosis, proliferation, oxidative stress, mitochondrial toxicity and alteration of calcium homeostasis.”
The limited availability and growth potential of primary mammalian cells reduce their potential for being used for HCS. However, target tissues/cells derived from human embryonic stem cells and induced pluripotent stem cells (iPSCs), can represent an extremely useful tool for drug screening, such as cardiomyocytes (CMs) or neurons which are actively used for drug screening.
A critical step in establishing a reproducible cell-based model for drug screening and/or disease modelling using iPSCs, is to ensure a reliable, scalable source of cells that will differentiate predictably and mimic human tissues in an artificial in vitro environment for HCS.

Di Marco describes her team’s experience with iPSCs in both 2D and 3D, with embryoid body formation used to differentiate the stem cells into the three main embryonic cell types: ectoderm, endoderm and mesoderm. “We have strong experience with neuronal differentiation and use these reproducible models specifically for Huntington disease research. Notably, iPSC-derived neurons obtained from Huntington disease patients harbour the CAG expansion in huntingtin (HTT) and represent a disease-relevant platform for drug testing, and to study the etiopathology of this neurodegenerative disease at the cellular and molecular level.”
In vitro model of the human Blood-Brain-Barrier (BBB)
For CNS drug discovery, Di Marco’s team established a human BBB model using endothelial cells derived from human iPSCs. Such models mimic major in vivo BBB characteristics including tight junctions
and transport mechanisms. This in vitro iPSC-hBBB model can be integrated in a screening paradigm of promising compounds for the treatment of brain diseases.
“Using normal or patient-derived iPSCs as a source of brain-like endothelial cells, we can create an in vitro model of the human BBB for mechanistic and functional studies to improve our knowledge of the BBB in both healthy and diseased states, applying a combination of HCS- and non-HCS-based assays,” states Di Marco. “Using an established step-wise approach, a working cell bank of endothelial progenitors is derived from iPSCs, stored in liquid nitrogen and can then be used in the final assay set-up all within one week, while also ensuring consistency and reproducibility.” The results of the researchers’ studies were published in Molecular Pharmaceutics(3) and Cells(4).
IRBM’s breadth and depth of experience in such areas as HCS and cell models, combined with IRBM’s passion for science, promotes a can-do, relentless approach to overcoming challenges, founded on integrity and paired with an impressive suite of capabilities under one roof that consistently delivers value to its partners worldwide.
References
1 Lang, P., Yeow, K., Nichols, A. et al. Cellular imaging in drug discovery. Nat Rev Drug Discov 5, 343–356 (2006).
2 Chandrasekaran, S.N., Ceulemans, H., Boyd, J.D. et al. Image-based profiling for drug discovery: due for a machine-learning upgrade?. Nat Rev Drug Discov 20, 145–159 (2021).
3 Application of an in Vitro Blood-Brain Barrier Model in the Selection of Experimental Drug Candidates for the Treatment of Huntington’s Disease. Di Marco A, Gonzalez Paz O, Fini I, Vignone D, Cellucci A, Battista MR, Auciello G, Orsatti L, Zini M, Monteagudo E, Khetarpal V, Rose M, Dominguez C, Herbst T, Toledo-Sherman L, Summa V, Muñoz-Sanjuán I. Mol Pharm. 16 (5):2069-2082. (2019)
4 Establishment of an in Vitro Human Blood-Brain Barrier Model Derived from Induced Pluripotent Stem Cells and Comparison to a Porcine Cell-Based System. Di Marco A, Vignone D, Gonzalez Paz O, Fini I, Battista MR, Cellucci A, Bracacel E, Auciello G, Veneziano M, Khetarpal V, Rose M, Rosa A, Gloaguen I, Monteagudo E, Herbst T, Dominguez C, Muñoz-Sanjuán I. Cells. 2020 Apr 16;9(4):994
IRBM is a contract research organization with an interdisciplinary team of over 200 scientists providing services across the entire preclinical drug discovery and development continuum. Based near Rome, Italy, the company collaborates with global organizations and has a track record of success in the development of novel therapeutic agents, including small molecules, peptides and monoclonal antibodies.
LEARN MORE @ www.irbm.com | [email protected]

