Scientists at the University of Exeter have developed a new technique that could help to reduce antibiotic prescribing by predicting within minutes which drugs may be effective against specific bacteria. The novel microfluidic technique works by exploiting the fluorescent properties of antibiotics to see if they are taken up by the bacteria and have a chance of being active. If the antibiotics can penetrate through the bacterial cell membrane and are taken up, then the bacteria glow brighter under the microscope.
The technology, which can quantify antibiotic accumulation at the single-cell level, is currently in the early stages of development, but the team hopes that the miniaturized devices could be used in clinical settings to help aid antibiotic prescribing, and also help scientists to develop more effective antibacterial compounds. Research lead Stefano Pagliara, PhD, a biophysicist at the University of Exeter Living Systems Institute, said, “We’re really excited about the potential for this technique to make a meaningful reduction in prescribing, helping to fight the global threat of antibiotic resistance. At the moment, it can take days for clinicians to get a lab result, which involves growing bacteria, but there is still some guesswork involved. Our technique could reduce the use of multiple antibiotics to try and fight a bacterial infection.”
Pagliara and colleagues report on their technology in Lab on a Chip, in a paper titled, “Single-cell microfluidics facilitates the rapid quantification of antibiotic accumulation in Gram-negative bacteria.” Antibiotic resistance is recognized as a major global health threat; it is predicted that by 2050, around 10 million people will die annually of microbial infections. “There is thus a desperate need to refresh the antibiotic development pipeline and develop new technologies to optimize antibiotic treatment,” the authors wrote.
Infections caused by Gram-negative bacteria are of particular concern, the team continued. These bacteria have a double-membrane cell envelope that represents what the Exeter researchers call “a formidable barrier” to antibiotic accumulation in the microorganisms. To get across the outer membrane, antibiotics have to penetrate through protein pores, or porins, which the bacteria use to take up nutrients. Gram-negative bacteria also have efflux mechanisms that effectively pump toxic compounds, such as antibiotics, back out of the cell. The activity of these porins and efflux pumps vary under different microenvironmental conditions, and also within bacteria that have the same genetic makeup that are exposed to similar environmental conditions. “Quantitative methods for studying drug accumulation in individual bacteria are therefore crucial for understanding this molecular transport landscape, to drive the rational development of the next generation of antibiotics,” the authors noted.
The most commonly used techniques for studying antibiotic accumulation in bacteria are population level assays, but these can’t evaluate drug accumulation at the single-cell level, the investigators stated. Current techniques also commonly rely on complex washing steps that can impact on cell physiology, or increase the likelihood of damaging the bacterial cells. “There is, therefore, a need to fundamentally change the experimental approach for quantifying antibiotic accumulation in bacteria, to incorporate single-cell level methodologies and the ability to study drug accumulation after exposure to different nutrient conditions or in different metabolic states,” they stated. Ideally, a workable approach should also be simple to carry out, so that it can be used by the industry to help antibiotic R&D, or in clinical settings.
The technique developed by the Exeter scientists combines time lapse autofluorescence microscopy with a microfluidics device, to investigate bacterial uptake of the antibiotic ofloxacin, which fluoresces under ultraviolet light. The bacteria glow when the antibiotic is taken up by the bacterial cells, and so has a chance of being effective against them. In contrast, if the antibiotic hasn’t been taken up, the bacteria don’t fluoresce.
The resulting system quantifies antibiotic accumulation in Gram-negative bacteria at the single-cell level. “Here, we address these myriad challenges by harnessing the power of microfluidics and auto-fluorescence microscopy to study the accumulation of the fluoroquinolone antibiotic ofloxacin in up to hundreds of individual bacteria confined in well-defined microenvironments,” the authors explained. “We reported, to the best of our knowledge, the first quantitative comparisons between drug accumulation in cells in different metabolic states and in cells with specific transport pathways disabled.
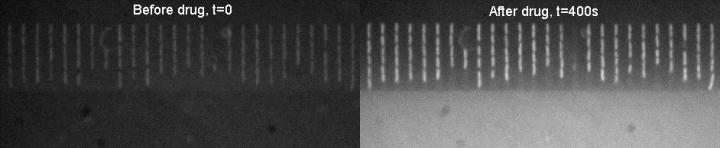
Moreover, they pointed out, unlike the majority of techniques that involve complex washing steps after drug delivery, or are limited to certain specific media conditions, their microfluidic platform makes it possible to study drug accumulation in different microenvironments and cellular metabolic states.
The team’s experimental results indicated that ofloxacin accumulates to a greater extent in growing, rather than stationary phase bacteria. While this may be due to differential expression of key membrane transport pathways, the scientists acknowledged that further investigation will be needed. “Future studies on the differences in accumulation between cells in different physiological states may also contribute towards a better understanding of antibiotic tolerance; indeed, it is known that growth conditions play an important role in the development of tolerance to various antibiotic classes,” they wrote. “Unlike traditional techniques, our assay is rapid, studying accumulation as the cells are dosed with the drug. This platform provides a powerful new tool for studying antibiotic accumulation in bacteria, which will be critical for the rational development of the next generation of antibiotics.”
Jehangir Cama, PhD, an industry research fellow at the Living Systems Institute, who performed the experimental work for the reported research, said: “Our next step is to further develop this exciting new method by combining it with more advanced microscopy techniques, to see where exactly the antibiotics go when they enter the bacteria.”
The authors concluded, “…our technology represents a step-change in the way drug accumulation in bacteria is studied, with potential benefits to the drug development and diagnostics industries, infectious disease specialists, as well as to the large academic community studying antibiotic transport in Gram-negative organisms…Besides its applications in drug development, our microfluidic platform can complement recently developed lab-on-chip antibiotic susceptibility testing systems to determine the contribution of reduced drug accumulation to bacterial survival in clinical settings.”
The team is now working on further expanding the technique, by manipulating the fluorescent qualities of other forms of antibiotics so that they can work in the same way. Further research in this area has been funded by QUEX, a partnership between the University of Exeter and the University of Queensland in Australia. The Queensland team, led by Mark Blaskovich, PhD, director of the Centre for Superbug Solutions at the Institute for Molecular Bioscience, is developing fluorescent versions of other antibiotics so they can be tested in a similar manner. Blaskovich added: “I am enthused about the opportunities to improve our fundamental understanding of the interactions between antibiotics and bacteria and how this leads to antimicrobial resistance, by combining our novel antibiotic-derived probes with the cutting-edge single cell analysis capabilities of the Exeter group.”


