Voltaire argued, “Perfection is attained by slow degrees; it requires the hand of time.” However, in drug discovery projects, perfection can’t be attained soon enough. A project to develop a perfect drug, or a drug that is as safe and effective as possible, may never begin if it can’t be completed quickly. Such predicaments must have been familiar to the philosopher. After all, he also complained that the perfect is the enemy of the good.
If only Voltaire were here today to see how drug discovery is being accelerated by new technologies. He might reverse himself and declare that the perfect can in fact be the friend of the good.
Several new technologies that are being used to accelerate drug discovery are highlighted in this article. They include small molecules that conditionally modulate proteins in their functional state; three-dimensional (3D) cell models, or organoid models, that assist with target identification, high-throughput drug screening, drug efficacy analyses, and toxicity studies; technologies that stabilize G protein–coupled receptors (GPCRs) to enable the discovery of agonistic biologics; and automated instrumentation that enables the overnight synthesis of mRNA.
Drugging the “undruggable”
“While about 50% of the proteome plays a role in human disease, only 10% is accessible to conventional therapeutics,” says Alexandra Glucksmann, PhD, president and CEO, Cedilla Therapeutics. To overcome that challenge, the company is pursuing a “target centric” approach to drug discovery. “We are developing small molecules that conditionally modulate proteins in their functional state,” Glucksmann explains. Such molecules, she continues, may provide “unprecedented precision therapeutics.”

According to Glucksmann, to access traditionally undruggable targets, it’s critical to understand the importance of form and context of targets in their natural settings: “We are bringing forward a new dimension in precision oncology by selectively inhibiting oncogenic drivers using small molecules that inhibit proteins in their functional states.”
Since much of the distinctiveness of proteins depends on post-translational modifications, Glucksmann suggests that when active sites are targeted, focusing on post-translational modifications can be effective. For example, solid tumors such as mesothelioma and certain types of squamous cell cancers feature a dysregulated key component of the Hippo signaling pathway called TEAD (transcriptional enhanced association domain). The aberrant regulation of this nuclear transcription factor is increasingly implicated in resistance to targeted therapies.

President and CEO, Cedilla Therapeutics
“The palmitoylation of TEAD is critical for its stability and function,” Glucksmann points out. “Through genetic screens, we identified a very selective molecule that inhibits the function of TEAD by binding to this specific pocket.”
The company is also targeting proteins that form complexes. “We are developing small molecules that selectively bind the cyclin-dependent kinase 2 (CDK2)/cyclin E complex,” Glucksmann elaborates. “We have developed an inhibitor with exquisite selectivity that is likely to translate into a better safety profile as compared to traditional kinase inhibitors.”
The key to the company’s approach is to find a validated target and develop drugs using a holistic approach. “[Such an approach] integrates matured technologies such as chemical biology, structural biology, cryo-electron microscopy, and proteomics,” Glucksmann notes. “This allows us to pursue targets that were deemed undruggable.”
While Cedilla is focused on precision oncology with a number of programs in the preclinical stage, Glucksmann feels that this strategy ultimately has many other applications. “This is not a shotgun approach,” she insists. “Rather, it is a target-centric pursuit.”
3D cellular models
Traditionally, once a potential drug candidate is identified, the compound is tested for efficacy and toxicity in two-dimensional (2D) cell cultures and animal models. According to Rosha Poudyal, PhD, science and technology advisor, 10x Genomics, 2D models present significant challenges for drug discovery. “They lack cellular heterogeneity, cell-to-cell interactions, and the ability to recapitulate the tissue microenvironment found in vivo,” she details. “Similarly, drugs that are successful in animal models can fail in the clinical phase because of reproducibility issues that are due to genetic differences between animal models and humans.”
Poudyal suggests that some of the challenges encountered with 2D models can be avoided if organoid models are used in concert with single-cell and spatial solutions. “Organoid models are tiny, self-organizing, in vitro 3D cellular models,” she explains. “They exhibit heterogeneous cellular composition and mimic the physiology of the organ in vivo, providing a platform to study human physiology that might be difficult to recapitulate in animal models. Organoids can be used for target identification, high-throughput drug screening, drug efficacy, and toxicity studies, and they have the potential to link preclinical and clinical data.”
When organoids are used in combination with single-cell technology to assess drug responses, researchers can assess cell-to-cell heterogeneity, understand gene regulatory networks, and perform pathway analysis. “Furthermore,” Poudyal adds, “single-cell resolution allows the identification of subpopulations of cells that are not as responsive or are resistant to the drug application.”
Poudyal says that these kinds of studies can be performed with patient-derived tumor organoids: “These models are gaining traction in precision medicine. They can be used in screening applications to identify drug(s) that would provide the most benefit to the individual patient. Similarly, neural organoids provide a platform to model neurodegenerative diseases, such as Alzheimer’s disease, where many therapies have been successful in animal models but have failed to get approved for clinical use.”
Recently, 10x Genomics launched Chromium X, a next-generation single-cell platform that can be used with the company’s new high-throughput assays to enable cost-effective, large-scale, single-cell experiments. The company offers many other technologies, including Visium, a spatial gene expression technology.
Functionally flexible GPCR drugs
GPCRs constitute the largest and most diverse set of membrane proteins in the body. Members of this receptor superfamily bind to a broad range of molecules and help direct many key physiological processes. “Because GPCRs are involved in various diseases, they account for about one-third of all market drugs,” says Christel Menet, PhD, CSO Confo Therapeutics. “But they still face many challenges such as lack of specificity and unwanted side effects.”
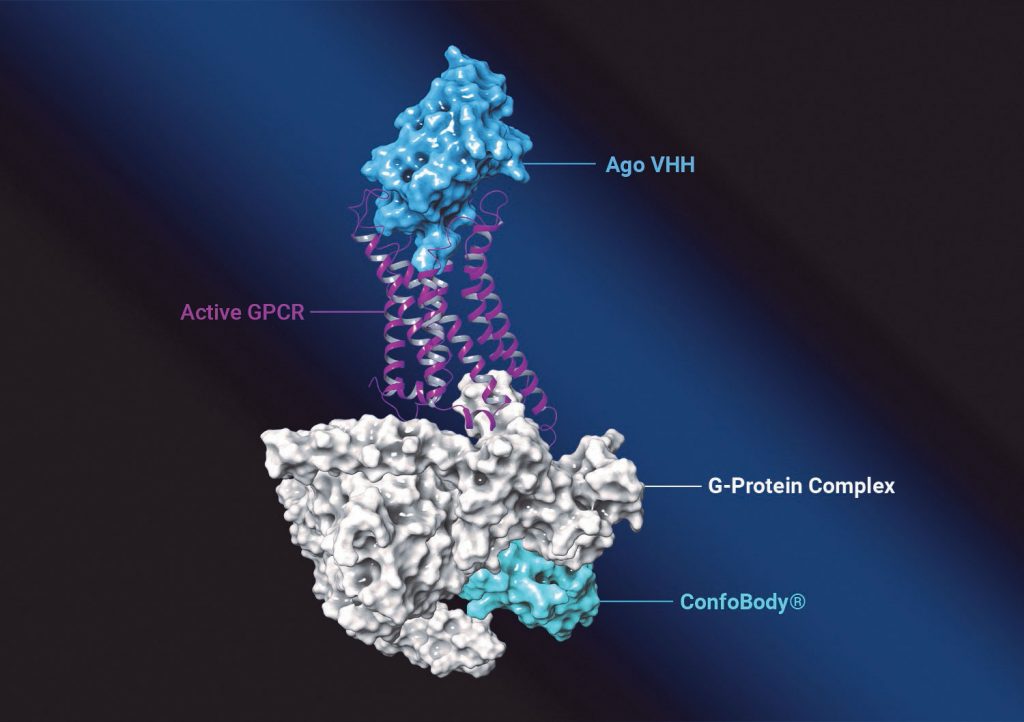
Menet advises that while therapeutic antibodies and other biologics have gained considerable attention with the launch of the first two GPCR-targeting antibodies (erenumab and mogamulizumab), the discovery of such antibodies remains challenging.
“As far as we are aware, all GPCR antibodies that are in development or on the market are antagonists,” she continues. “Confo has demonstrated that, in addition to structure-based drug discovery of small molecules, its unique technology to stabilize the GPCR in its native conformation and natural environment can also be leveraged to discover agonistic biologics with very high potency and selectivity.”
To stabilize GPCRs, the company uses ConfoBodies, single-domain antibodies specific for a particular target conformation. They are generated by state-of-the-art VHH (single domain antibody fragment) discovery methods, either from immune or non-immune libraries. Menet elaborates, “We also recently discovered more universal methods to applying the Confo technology without the need for an additional ConfoBody discovery for every new GPCR target we want to drug. This of course drastically improves the efficiency and success rate of our approach.”
Once Confo scientists are happy with the pharmacological profile of the ConfoBody, they use the ConfoBody-stabilized GPCR as the driver for their drug discovery process for chemical and/or biologics approaches. “The resulting therapeutic candidates,” Menet explains, “are then characterized in a cascade of different in vitro and in vivo models to examine their relevance for treating the disease(s) of interest.”
The company is building its proprietary pipeline and expanding its outlook. “We would like to be able to pursue any therapeutically relevant GPCR, including less-well-characterized and orphan GPCRs,” Menet comments. “Ideally, we would also expand beyond GPCRs to other classes of membrane proteins.”
Digital info in, mRNA out
Vaccine and therapeutic researchers are increasingly focusing on mRNA-based platforms for discovery, especially in light of the success of the mRNA-based COVID-19 vaccines. “Typically, mRNA discovery workflows begin with an iterative design-build-test process that requires researchers to design their candidate mRNA molecules and synthesize them prior to testing or screening for promising lead molecules,” observes Jyotsna Venugopal, PhD, director of product marketing at Codex DNA. “This process can be confoundingly empirical. It can take multiple iterative cycles before lead molecules are identified.”
To address this challenge, Codex DNA has developed a novel mRNA solution. “Our automated benchtop synthetic biology workstation—the BioXp 3250 system—enables hands-free synthesis of mRNA in a single overnight run,” Venugopal asserts. “This empowers researchers to avoid the weeks or months of wait time and challenges associated with acquiring synthetic mRNA and thus accelerates the evaluation of candidate molecules in their discovery process.”
The instrumentation performs all the steps required to go from DNA oligos to synthesized, assay-ready mRNA in a single automated run. According to Venugopal, accomplishing all these steps in a typical laboratory can require multiple scientists, numerous steps, varied skill sets, and long lead times. “In contrast,” she says, “the BioXp 3250 can produce up to 16 synthetic mRNAs in a single automated, overnight run.”
Venugopal also notes the importance of cooperative efforts: “We have collaboration agreements with various industry partners and believe this is important to advancing our mRNA workflow solutions for key applications.”
PROTACs and TUBEs
A promising drug discovery approach is targeted protein degradation. To date, this approach has emphasized the development of small-molecule drugs called PROteolytic TArgeting Chimeras (PROTACs). These “degrader” drugs work by targeting the ubiquitin-proteosome system (UPS).
PROTACs are heterobifunctional molecules that consist of a ubiquitin-ligase-targeting ligand, a chemical linker, and a distinct protein-binding ligand. The complex promotes protein degradation via production of polyubiquitin chains on the protein of interest.
“The major problem in PROTAC drug discovery is slow PROTAC design, synthesis, and testing,” observes Karteek Kadimisetty, PhD, director of R&D at LifeSensors. “The PROTAC community needs a fast method for monitoring PROTAC function and generating reliable data. LifeSensors has translated the traditional western blot assay into a faster method by monitoring PROTAC-mediated ubiquitination of target proteins on a microtiter plate–based assay called PROTAC Plate.”
PROTAC discovery is also hampered by a large gap in understanding the fine link between ubiquitination and degradation. “Current methods to quantify this process are of limited utility in drug discovery,” Kadimisetty points out. “We addressed these gaps utilizing Tandem Ubiquitin Binding Entities (TUBES) in high-throughput biochemical and cell-based assays.”
TUBEs are specifically designed to isolate polyubiquitinated proteins from cell lysates and tissues. Kadimesetty explains, “These high-affinity capture reagents bind polyubiquitin chains. One can perform quantitative pulldown of ubiquitinated proteins and establish correlations between PROTAC-mediated ubiquitination and degradation of proteins. Further, mass spectrometry ubiquitin proteomics has facilitated PROTAC-mediated ubiquitination of target proteins. These and other methods have clarified PROTAC mechanisms of action.”
“TUBEs will help cell and molecular biologists better understand the role of novel polyubiquitin chains,” Kadimisetty predicts. “Further, if we understand the role of other polyubiquitin chains and E3 ligases responsible for novel functions, new nondegradative PROTACs can be designed to broaden their therapeutic potential.”


