When we say it’s the little things that count, we seldom consider whether those things could include microbes. We’re too aware of microbes like flesh-eating bacteria, respiratory viruses, and other pathogens. But not all microbes are sources of invariable dread. Many microbes are innocuous or even beneficial. And some go “bad” only if they are in a microbial community that is falling into dysbiosis—that is, losing its health-sustaining balance.
We should remember the “good” microbes, which number in the trillions and effectively serve as our second genome. This isn’t just a matter of fairness. It’s about our own self-interest. An open-minded view toward microbes will help us see the qualitative and quantitative fluctuations that occur in our microbial communities—fluctuations that can make the difference between health and sickness.
A healthy curiosity about the ups and downs of microbial communities was clearly evident at the 8th Annual Translational Microbiome Conference. This event, which was held in Washington, DC, last April, featured the latest microbiome research and advice on how to achieve commercial and clinical success. Key discussions from the event are revisited in this article.
Topics covered here include the use of bioinformatic tools to streamline the development of live bacterial products (LBPs) such as “living drugs”; the recognition of gut microbiome patterns to create an index for predicting health conditions; the use of metatranscriptome analyses and metagenome assembly algorithms to characterize 16S rRNA microbial genes; the development of therapeutics that incorporate recombinant probiotics; and the selection of LBP strains to reprogram the infant gut microbiome to a healthy state.
LBP discovery services
Finding candidate strains for LBPs can be a time consuming and expensive process. To facilitate this process, Clinical Microbiomics, a contract research organization, offers a range of microbiome analysis services.

Clinical Microbiomics
“LBP discovery starts by studying microorganism properties preclinically for key genotypic and phenotypic traits,” says Heidi Hau, PhD, the company’s vice president, for scientific affairs and partnerships. “LBP development includes the identification, isolation, and characterization of microorganisms that make up the drug product.
“The process typically begins by isolating and screening a large number of candidate strains to identify those with a favorable safety profile that are anticipated to provide the desired properties for the intended therapeutic benefit. Screens may include in vitro assays, ex vivo systems, preclinical animal studies, and finally, human clinical trials. Further, strain failure rate can be high when advancing to scaled fermentation or animal studies, despite extensive screening.”
According to Hau, there is a need for methods that characterize strains for desired genotypes and phenotypes, and that highlight unique attributes facilitating strain selection before advancing to assay-based characterization and animal models. To solve these challenges, the company developed a data-driven approach.
“Clonal-level phylogenetic microbiome profiling, which shows how closely related a candidate strain is to other known strains, forms the basis of our platform,” Hau relates. “This is combined with comparative genomics analysis and a data-driven platform that links known desirable (or undesirable) phenotypes, like engraftment potential, optimal growth conditions, associated clinical outcomes, and so on, to microbial populations.
“Integrative analyses that combine phylogeny, genotype, phenotype, and outcome data can be used to narrow and de-risk strain selection prior to assay-based characterizations. This translates to significant cost and time reductions from discovery to preclinical validation. Moreover, mechanism of action can be hypothesized through this approach as well as discovery of microbiome-derived small molecules for specific application areas.”
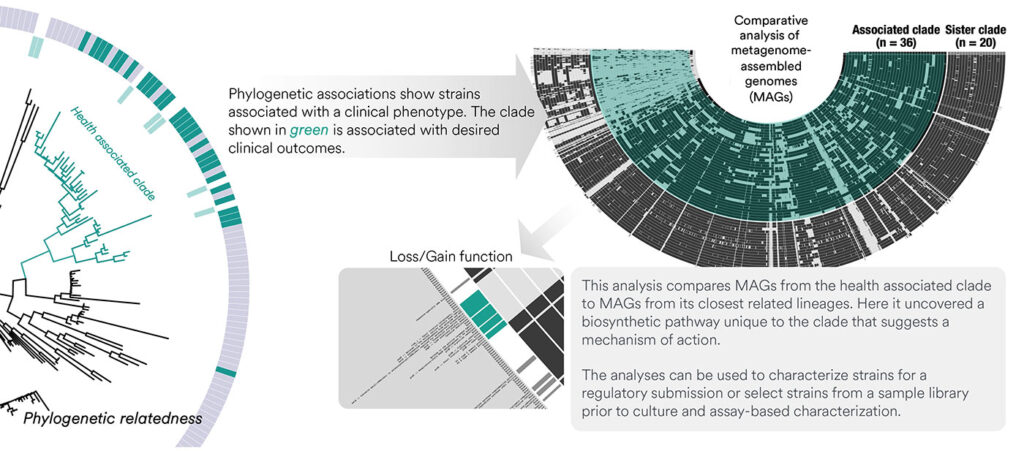
Hau suggests that Clinical Microbiomics is the only contract research organization that leverages clonal-level resolution to facilitate engraftment analysis and deep comparative genomics. “We are pioneers in the microbiome field,” she asserts. “We have generated over 225 published articles—over 25 of which have appeared in Nature, Cell, or Science journals.”
The gut microbiome index
Of the trillions of microbes that inhabit the body, 95% reside in the large intestines. “These are mostly beneficial,” says Nur A. Hasan, PhD, CEO, EzBiome. “They help digest our food, produce vitamins, prevent pathogen colonization, and produce metabolites that interact and regulate our immune, metabolic, and nervous systems. They form an ecosystem. It is called the gut microbiome, and it can influence our health and wellbeing.”

EzBiome
Hasan says that unfortunately there is as yet no reliable measure that can accurately reflect the current state of gut microbiome health. “A robust gut microbiome-based health index would offer an era of preventive medicine not only by measuring the level of any imbalances, but also by predicting the underlying cause of such imbalances to enable potential health-restoring solutions.”
To help meet these challenges, EzBiome and its parent company CJ Bioscience developed the Precision Taxonomy discovery platform. The companies soon put Precision Taxonomy to good use. In a study led by Jon Jongsik Chun, PhD, a professor of bacteriology and bioinformatics at Seoul National University and the CEO of CJ Bioscience, the companies used their platform and expert-curated reference databases to better characterize the gut microbiome data of a large, multiethnic population.
The study assessed data from about 120,000 individuals and encompassed host covariates such as health status, lifestyle, diet, medication, and bowel habits. Members of the study population included healthy people and people who were suffering from one or more diseases. (More than 25 diseases were recorded.)
“This work comprises datasets collected and stratified from various public repositories, as well as high-quality datasets collected and generated by our group,” Hasan notes. “We used a standardized sample-to-result workflow through joint research with 14 hospitals and medical research institutes over six years.”

Using the microbiome patterns observed in healthy and nonhealthy individuals, the companies then devised an index that could consistently predict health conditions from human gut microbiome data. The companies called it the gut microbiome index (GMI).
“Compared to ecological metrics, the microbial marker index, and machine learning approaches, GMI distinguished between healthy and nonhealthy individuals with a higher accuracy across various datasets,” Hasan asserts. “Therefore, we propose GMI as a potential index to measure the health status of the gut microbiome. This has been verified from multiethnic data of various diseases and a two-year citizen science project that Chun and his team carried out.”
The companies will continue to collect geographically diverse sociotemporal and multiethnic datasets at a global scale to expand and further improve their proposed GMI index.
Beyond DNA and microbial metagenomics
Researchers studying the microbiome must decide which kinds of data are best suited for their studies. Options include genomics, transcriptomics, and metabolomics data. For a perspective on these options, we spoke with Brice Le François, PhD, senior scientist, DNA Genotek and senior R&D manager, technology, Diversigen. (DNA Genotek provides sample collection devices that preserve host and microbial analytes. Diversigen offers microbiome services.)

DNA Genotek, Diversigen
“Most microbiome studies tend to focus on DNA-based sequencing approaches (metagenomics) to understand the role of microbes in human health and disease,” Le François says. “DNA is very useful to establish which organisms are present in a sample and infer their functional potential, but it fails to identify the species that are active and their functional outputs in vivo.”
“Past studies used metatranscriptomics to interrogate the gut microbiome, but they lacked optimal tools and focused on healthy and underpowered cohorts, limiting interpretations that could be drawn,” Le François continues. “New sample collection, library preparation, and data analysis technologies developed in the past few years have made metatranscriptomics more achievable than ever.”
Le François and his team performed a pilot study to compare the response of gut bacteria to human intermittent fasting at the metagenomics versus metatranscriptomics levels to better understand the value each analyte provides. “Our study shows that gut metatranscriptomics captures dynamic gene expression changes that correlate with intermittent fasting. In contrast, gut metagenomics profiles tend to be highly stable,” Le François asserts. “Our results make a strong case for metatranscriptomics being more suited to studying the response of the microbiome to changes in its environment.”
High-resolution profiles
One of the key ways to identify and characterize different microbial taxonomies in complex mixtures is to sequence the 16S rRNA genes. For example, the prokaryotic 16S rRNA gene consists of about 1,500 base pairs encompassing nine variable regions interspersed among conserved regions.

Pacific Biosciences
“All bacteria and archaea species contain a 16S gene, and the variation within this phylogenetically important gene enables taxon identification,” says Jeremy Wilkinson, PhD, senior staff specialist, microbial genomics, Pacific Biosciences (PacBio). “It’s also a less expensive way to capture taxonomic profiles than shotgun metagenome profiling. Amplicon-based approaches, like 16S, are also important for largely host-contaminated sample types (such as tissue and blood) and low-microbial-biomass sample types (such as swabs).”
Wilkinson suggests that researchers can use a PacBio technology called HiFi sequencing to capture full-length 16S rRNA. “In the past several years,” he asserts, “this technology has enabled researchers to profile microbiomes with significantly higher resolution at the species and strain level with as much as 99% of reads classified to the species level.”
Most current technologies cannot produce a high percentage of classified reads for accurate species identification. PacBio has developed workflows for full-length 16S profiling, shotgun metagenome taxonomic and functional profiling, and shotgun metagenome assemblies.
“With the development of new metagenome assembly algorithms for HiFi reads (including hifiasm-meta), it’s now possible to reconstruct full metagenome-assembled genomes (MAGs) for high-abundance species,” Wilkinson elaborates. “However, discontiguous assemblies occur for lower-abundance taxa. Post-assembly binning methods are therefore required to identify these.
“We have developed the HiFi-MAG-Pipeline that automates major steps, including a custom binning strategy tailored to long-read assemblies, quality filtering, and taxonomic identification. We show that complete, circular MAGs are routinely produced and that the HiFi-MAG-Pipeline recovers more high-quality MAGs than other methods.”
Oral microbiome immunotherapy
More than 1.6 million adults and children suffer from inflammatory bowel disease, which may manifest as Crohn’s disease or ulcerative colitis. Since IBD features a breakdown of intestinal immune regulatory signals, it is most commonly treated with systemic immunosuppressants or cytokine blockers, both of which can cause serious side effects.

Rise Therapeutics
An alternative approach is proposed by Christian Freguia, PhD, senior vice president of research, Rise Therapeutics. An oral microbiome immunotherapy, he says, could harness “the ability of human commensal bacteria to skew the immune response and rebalance the human immune system.”
Rise is targeting inflammatory bowel disease with an oral microbiome immunotherapy called R-3750. Freguia points out that R-3750 is a synthetic biology medicine that consists of a “safe probiotic engineered to deliver an immunotherapeutic protein called surface layer protein A (SlpA).” He adds that R-3750 represents a “precision targeted microbiome product encompassing a safe delivery vehicle and a novel protein that is key in balancing the immune system.”
R-3750 (SlpA) engages a specific receptor expressed on dendritic cells lining the gut endothelium to reestablish colonic immune homeostasis by reducing inflammation, improving gut membrane barrier integrity, and reestablishing a normal microbiome composition. “The recombinant probiotic is encapsulated for easy oral administration,” Freguia notes.
The company is evaluating R-3750 in a Phase I study for ulcerative colitis. The company’s pipeline also consists of additional oral precision synthetic biology–based immunotherapeutic products.

Reprogramming unhealthy infant microbiomes
“We are developing live bacterial products that get at the root cause of disease,” states Nikole Kimes, PhD, CEO and co-founder, Siolta Therapeutics. Siolta (pronounced sheel-ta) is an Irish Gaelic word for “seeds” and signifies re-seeding the gut microbiome with naturally occurring healthy bacteria. She continues, “We are using a targeted approach to microbiome-based interventions. Some companies utilize fecal materials as therapeutics, and although they may show efficacy, these materials are very undefined.” It is unclear which organisms are involved, or how their influences can be explained.

Siolta Therapeutics
Kimes says that Siolta always starts with clinical data from humans. “The data tells us a story of what is different and misaligned and how it may promote disease,” she explains. “Then we go into the laboratory to identify which functions are synergistic between different organisms. This instructs us how to replenish with organisms that provide the function needed.”
Siolta identified a microbial signature linked to metabolic and immunological functions that drive disease in a certain population of children. To treat the children in this population, the company is developing a product that can reprogram their microbiomes. “Reprogramming results in microbiomes that look more like those in healthy infants,” Kimes says, “and it prevents the immunologic cascade that leads to disease downstream.”
The lead product, STMC-103H, is currently undergoing a Phase II proof-of-concept trial in newborns for the prevention of atopic dermatitis. It is called the Allergic Disease Onset Prevention Study (“ADORED”). The LBP treatment is administered as a lyophilized powder. Siolta also has a preclinical program developing a treatment for bacterial vaginosis which impacts women and their ability to have healthy pregnancies.
“It is clear the microbiome is driving many aspects of our health,” Kimes declares. “Branching out and considering the systemic impact of the microbiome is going to allow us to approach a plethora of diseases that have not been touchable to date.” She is also optimistic about the future of Siolta’s specially selected LBPs: “We are excited about utilizing our defined LBPs not only to treat disease, but also to prevent it. That’s a game changer in healthcare.”