Sponsored content brought to you by

In biopharmaceutical manufacturing, the development of an expression system that stably produces high levels of desired recombinant protein can be a time consuming and resource-intensive process. In brief, this process requires a vector containing the geneselect s) of interest to be transfected into cells, and high performing clones to be identified and isolated. This process takes many months, requiring large amounts of personnel time, supplies and equipment. Our goal in this study was to modify a well-defined cell line development system to generate high performing recombinants in a time- and resource-efficient manner.
The CHOZN® platform
CHO select Chinese Hamster Ovary) cell lines are the preferred expression systems for the production of therapeutic proteins. MilliporeSigma has developed the CHOZN® platform, an off-the-shelf, integrated CHO cell expression system. Consisting of a rationally engineered cell line, paired cell culture media and feeds as well as the user-friendly CHOZN® Omics Explorer, the CHOZN® platform allows for selection and easy scale-up of high-producing clones for predictable production of biomolecules such as monoclonal antibodies and other recombinant proteins. Engineered with the CompoZr® Zinc Finger Nuclease select ZFN) technology, the key to the CHOZN® platform is the CHOZN® GS-/- cell line that is auxotrophic for the amino acid L-glutamine due to elimination of endogenous glutamine synthetase select GS). This glutamine auxotroph cell line is designed to be used with an optimized expression vector that contains the GS gene and two multiple cloning sites with linked CMV promoter-driven expression cassettes.
UCOE® technology
One of the most common problems encountered during the characterization of the biomanufacturing cell lines is poor expression stability of the recombinant therapeutic protein. Although there are many factors that can influence production and stability, one of the most well-studied and characterized is chromatin state surrounding the integrated transgene. In their native state, chromosomes can be tightly condensed into heterochromatin. This tightly wound form of DNA is not a good substrate for transcription enzymes, rendering it transcriptionally inactive. With most conventional cell line development vector systems, the integration of the transgene occurs in a random fashion with transgene recombination into either transcriptionally inactive heterochromatin regions or into transcriptionally active open euchromatin structures. These ‘position effects’ pose a significant burden to cell line development workflows, as many clones must be screened in order to identify clones bearing a transgene integrated in a locus supporting high levels of expression. Moreover, even when inserted into transcriptionally permissive open chromatin structures, these gene insertions can still be silenced via promoter DNA methylation. It is noteworthy that the most common heterologous promoter used in commercial systems today, the Cytomegalovirus select CMV) Immediate Early Promoter, is among the most sensitive to methylation and silencing. We queried whether there was a way to maintain chromatin in an “open” configuration and retain transcriptional activity which could increase the fraction of high-expressing clones and reduce the screening burden. Dr. Michael Antoniou at King’s College London discovered a method to bypass issues of silencing and promoter methylation select Neville JJ et al. Biotechnol. Adv. 35:557-564, 2017). He found patches of DNA that conferred expression stability to CMV promoter-driven genes when positioned 5’ of the CMV promoter.

Figure 1: UCOE®technology
The elements discovered by Dr. Antoniou and coworkers are called Ubiquitous Chromatin Opening Elements or UCOE®s. UCOE® sequences are methylation-free, GC-rich DNA sequences derived from housekeeping genes. In cells, housekeeping genes produce proteins that are required for basic cellular functions and thus transcription is maintained at a steady state by keeping the associated chromatin organized in an open, transcriptionally active configuration. The UCOE® sequences are easy-to manipulate genetic regulatory elements that maintain the neighboring DNA in an open orientation and allow stable expression of Cis-linked genes in recombinant cell lines. They not only overcome transgene silencing but also mitigate the “position effects” of the site of integration. As a result, UCOE® sequences increase the number of stably producing recombinants in a transfection. UCOE® technology can be used in mammalian cell lines to stably express a variety of therapeutic proteins such as antibodies, hormones, and blood factors on industry standard platforms select Figure 1).
Combination of UCOE® technology with the CHOZN® platform
In our search for improvements to our already robust CHOZN® platform, we investigated whether the addition of UCOE® technology would support its high performance with a shorter time to result. Since UCOE® technology increases the number of stably expressing recombinants in a transfection, we attempted to clone UCOE®/CHOZN® recombinants using the bulk pool approach and compared the results to our standard CHOZN® platform utilizing the minipool approach. Using the bulk pool approach could save up to 6 weeks of development time.
DNA vector cloning
The standard CHOZN® GS-/- vector containing both heavy and light chains of a model monoclonal antibody was used to demonstrate the current control platform select Control). The heavy and light chains are under the control of CMV promoters with introns and poly A sites. In order to assess the effects of UCOE® elements on stable recombinant protein production, two different UCOE® sequences were cloned 5’ to the CMV promoters of both light and heavy chain gene expression cassettes to generate the UCOE® vector. select Figure 2) Expression of each of the antibody chains was influenced by a different UCOE® sequence. The 3.0 kb UCOE® sequence was inserted 5’ of the light chain promoter and the 1.5kb UCOE® sequence was inserted 5’ of the heavy chain promoter.

Cell line development workflowselect s):
Cell line development begins with transfection of plasmid DNA into CHOZN® GS cells. This can be accomplished by electroporation, transfection with cationic lipids or other methods. Following transfection, the standard CHOZN® platform utilizes a minipool screening approach for generation of recombinants. The minipool method utilizes selection of a small number of transfected cells in small volumes. This permits high-producing recombinants to expand without competition from fast-growing low-producing recombinants. While this method is useful for picking rare, high-producing recombinants, the time required to complete selection and screening is 7-9 weeks, and this method also requires significant resources to complete the necessary screening steps select Figure 3). Since UCOE® technology overcomes pitfalls in generating high-expressing clones by enabling active, stable transcription, we hypothesized that we could bypass the minipool step and select clones by the more rapid bulk pool approach. The bulk pool approach utilizes selection and screening of a large number of transfected cells and is complete in 2-4 weeks. This method may select against less frequent, slow-growing, high-producing recombinants, and as a result, this method is not frequently used without enrichment steps which usually are time consuming and resource intensive. However, since UCOE® technology has been shown to increase the population of high producing recombinants, we hypothesized that we would be able to isolate high-expressing clones from bulk pools made with UCOE® technology. After high performing bulk- or minipools are selected, they are subjected to limiting dilution, single cell cloning. Following growth, clones are evaluated for titer. This process requires an additional 7-9 weeks of development time regardless of initial screening mode.

Results
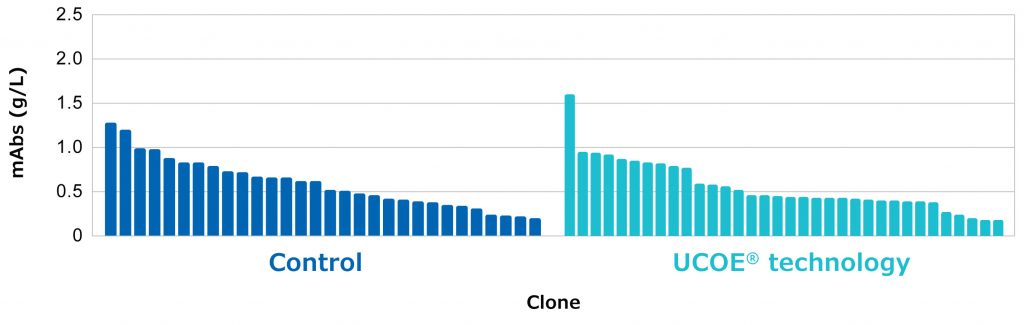
Approximately 300 clones were isolated for the Control vector using the minipool approach and for the UCOE® vector using the bulk pool approach. Clones were then scaled up and screened in batch for titer, and the top expressing clones were evaluated in a fed batch assay using EX-CELL® Advanced cell culture media and feeds. Titer was determined on day 14 of the fed batch assay select Figure 4). Control clones expressed a maximum of 1.3 g/L in small scale. This titer is within the expected range of the platform for different antibody molecules in a non-optimized fed-batch assay. By contrast, the highest expressing clone derived from the UCOE® vector using the bulk pool approach produced over 1.5 g/L. Since the titer of the UCOE® clone derived from bulk pools has similar performance to Control clones derived from minipools, it appears that UCOE® technology can be used to improve the CHOZN® platform by reducing cell line development resources and timelines.
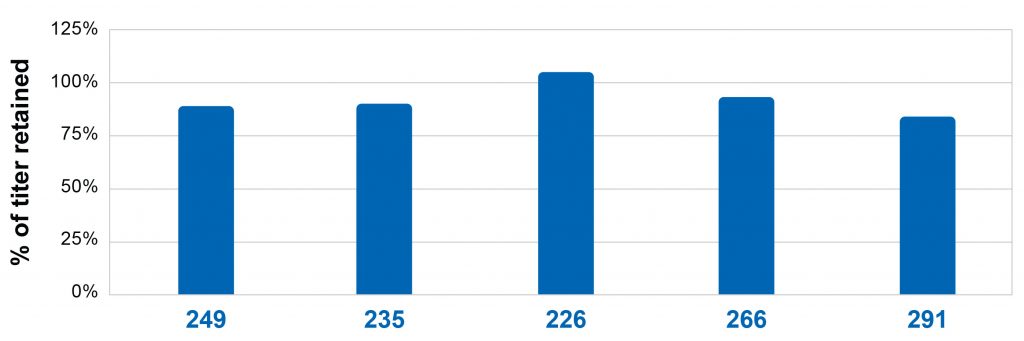
Stability of titer is maintained
In our study of the maintenance of titer stability, we took the highest expressing five CHOZN® GS-/- clones containing UCOE® sequences and made cell banks. One tube of the bank was thawed and passaged twice a week for 90 generations. Approximately every two weeks, new cell banks of the passaged cells were made. After 90 generations, banks on days 0 and 90 were thawed, permitted to recover and used to set up standard fed batch assays in EX-CELL® Advanced™ cell culture media and feeds. After 14 days, the titers of the cells were determined by HPLC. The titer of the 90-generation sample was compared to the titer of the day 0 bank and a ratio documented select Figure 5). All 5 clones maintained at least 75% of titer after 90 generations. These results demonstrate that protein expression of the top 5 clones is stable.
Conclusion
The combination of the CHOZN® platform with UCOE® technology enables rapid and robust cell line development. Using UCOE® technology in the CHOZN® platform, we were able to isolate clones with equal or better performance compared to the standard CHOZN® platform. In bulk pool mode, the combined technologies offer advantages for monoclonal antibody production, namely reduced resources and improved timelines. Since the impediments we have been trying to overcome include the longer timeline to results and consequent resource requirements, the ability to obtain sufficient antibody production from the bulk pool method with its shortened development time provides an option in streamlining and improving the cell line development for monoclonal antibody manufacturing.
Joe S. Orlando, PhD, is principal scientist and Kimberly Mann is senior scientist at MilliporeSigma.
© 2019 Merck KGaA, Darmstadt, Germany and/or its affiliates. All Rights Reserved. MilliporeSigma, the vibrant M, CHOZN, UCOE, CompoZr, and EX-CELL are trademarks of Merck KGaA, Darmstadt, Germany or its affiliates. All other trademarks are the property of their respective owners. Detailed information on trademarks is available via publicly accessible resources.
Related Content:


