April 1, 2006 (Vol. 26, No. 7)
Sample Collection and Prep Bottlenecks Combated with FTA Elute Matrix Devices
The National Center for Biotechnology Information determined that in 1994 there were over 2.2 million cases of adverse drug reactions in the U.S. Adverse drug reactions, which are rooted in individuals’ variable responses to medication, are a major cause of hospitalizations. Medications and doses, which are developed around the average response of a population, may affect each individual very differently.
Pharmacogenomics refers to the study of the numerous genes in an individual that determine his or her reaction to drugs. Each drug acts on a particular metabolic pathway in the human body. Each pathway involves many proteins, and therefore many genes.
A person’s genetic make up can have a profound influence on how he or she will respond to a drug. This discovery has led to the study of pharmacogenetics, which refers to the relationship between an individual’s genetic profile and his or her corresponding drug metabolism and response. Genetic tests, which analyze an individual’s genomic pattern of SNPs, can reliably predict his or her likely response to a given drug. The successful pharmacogenetic analysis of an individual will, ideally, lead to a doctor prescribing the best drug at a dose that will maximize the benefits while minimizing the risk.
Genetic testing facilities currently use several methods of genotyping, from traditional DNA sequencing to mass spectrometry to microarray technology. The first bottleneck of pharmacogenetics, however, is sample collection and DNA preparation.
The dominant method of sample collection involves the use of traditional vacutainer-based blood samples, which has a number of drawbacks. First, individuals participating in a study or drug trial must have their blood drawn by skilled personnel at a centralized location, and the drawn samples necessitate refrigeration. Shipping the blood, which is designated as a biohazardous material, requires ice and insulated packaging.
Extracting DNA from the blood samples, a task performed at a genotyping laboratory, requires labor-intensive and time-consuming methods. Finally, the extracted DNA samples must be stored in -70C freezers, which are both expensive and susceptible to electrical power outages. A common technique used to mitigate this risk involves storing portions of each sample in several locations, thus requiring redundant freezer units, labor, and electricity.

Fig.1: DNA samples are applied to the FTA Elute matrix where DNA is captured and protected. A hot-water elution method recovers pure DNA in solution in 30 minutes for genotyping studies.
FTA Elute Matrix Devices
The pressing need for a method and device to rapidly collect, ship, store, and purify DNA has led to the development of FTA Elute matrix devices from Whatma(www.whatman.com). FTA Elute is a chemically coated matrix that reversibly traps DNA from multiple types of biological samples. The chemicals within the matrix preserve DNA such that the samples are stable at room temperature for extended periods of time, which simplifies sample storage and shipping. Potentially harmful viruses and pathogens are inactivated by the matrix coating so that the samples are safe to handle and free of biohazardous material.
Just a few drops of blood from a finger-prick (approximately 10-40 µl) provide enough DNA for many genetic analyses. When used in combination with whole genome amplification (WGA) technologies, FTA Elute supplies enough DNA for a wide number of tests. Valuable biological samples can be archived or banked at ambient temperature, replacing expensive and space-consuming freezer banks.
FTA Elute matrix devices are a simple and rapid method for purifying DNA and can easily be automated for high-throughput genotyping laboratories, saving time and money. To purify DNA from FTA Elute, a small (3-mm) punch of the FTA card is placed in a microcentrifuge tube or multiwell plate and rinsed with water (Figure 1). The wash is removed, fresh elution water is added, and the disk is heated for 30 minutes at 95C. This step releases DNA from the matrix while proteins, impurities, and inhibitors remain bound to the matrix. The DNA is now ready for use in a number of genotyping methodologies.
Real-Time PCR
DNA eluted from the FTA Elute matrix device was subjected to quantitative PCR to demonstrate the quality of the purified DNA. Blood samples were collected from 10 separate individuals onto the FTA Elute matrix. Disks (3.0 mm) were extracted according to the water-extraction protocol in a final volume of 100 µl. Approximately 2.5 µl of the purified DNA was added to the real-time PCR mixture and amplified using the Yo-Pro 1 DNA binding dye (Invitrogen; (www.invitrogen.com). Figure 2 shows a tight grouping of curves with an average Ct of 26.58, which equals 22.14 ng of DNA in the 100-µl final volume. The yield of DNA from FTA Elute is sample-dependent. For blood and buccal samples, the yields are 55-70% and 60-75%, respectively.
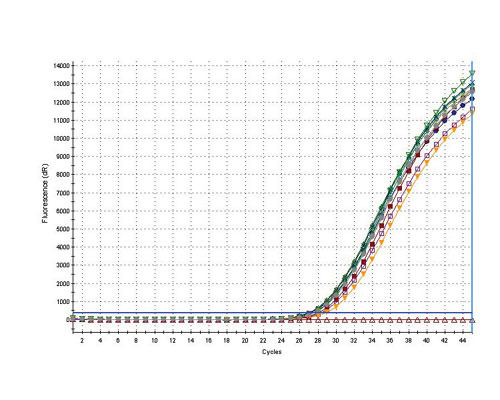
Fig.2: For this assay 2.5 mL of the extracted DNA was mixed with the DNA binding dye Yo-Pro 1 to monitor real-time PCR amplification. A no-template control was included in the assay and appears as the flat line at the bottom of the profile.
DNA Sequencing
DNA sequencing is the gold standard for detecting polymorphisms in gene sequences. DNA purified with FTA Elute was tested as a template for sequencing a 1.05 kb fragment from exon 1 of the 2B15 variant of the UDP-glucuronosyltransferase (UGT) gene. The G/T polymorphism at the D85Y locus was the target SNP. Figure 3 shows an example of three sequences from individuals showing polymorphisms in the sequence. This profile indicates that the DNA extracted with FTA Elute yields excellent sequencing data.
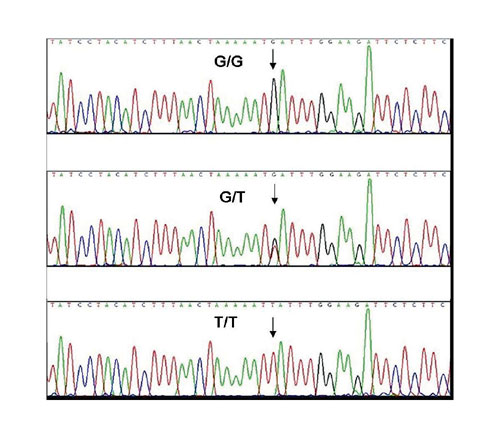
Fig.3: DNA samples from FTA Elute were used to amplify a 1.05 kb exon 1 fragment of the UCT2815 gene. The PCR product was treated with ExoSAP-IT(USB). Sequencing reactions were then run on an ABI Prism(r) 3100 instrument.
Multiplex Gene Deletion Assay
Many labs perform multiplex gene deletion assays using end-point PCR and agarose gel electrophoresis as a means of genotyping. Figure 4 shows a multiplex PCR performed with DNA purified with FTA Elute detecting a deletion in the UCT2B17 gene. The wild type and mutant genes are demonstrated by bands of 316 bp and 884 bp, respectively. As can be seen, the individual in lane 7 is homozygous for the mutant form of the gene. The ability to multiplex with multiple primers requires high-quality DNA to prevent mis-priming and nonspecific banding patterns. DNA purified by FTA Elute is robust and less than 1 ng is required to yield clean strong bands in the multiplex PCR.
FTA Elute serves both the pharmacogenomics and bio-banking markets. FTA Elute implements a simple, one-step method to collect and stabilize DNA for genotyping and SNP analysis. A quick, hot-water elution method yields high-quality DNA for a variety of genotyping methods such as Amplification Refractory Mutation Systems (ARMS) Scorpion, and Allele-Specific Oligonucleotide genotyping.
DNA from FTA Elute can also serve as a template for whole genome amplification, thus supplying a nearly unlimited amount of DNA for extended genetic studies. The archiving ability of the FTA Elute matrix is beneficial in “look-back“ studies and for archiving samples for future genetic analysis.


