October 15, 2012 (Vol. 32, No. 18)
Bead-Beating Technology Can Be Used to Disrupt or Homogenize Wide Range of Samples
Determining the internal pathogen burden of tissues and animals is an important component of infectious disease research. Yet the disruption of these tissues in order to release an intact, viable pathogen for quantification or further experimentation is problematic.
The Precellys®24 homogenizer (Figure 1) from Bertin Technologies is an established method based on bead-beating technology to disrupt or homogenize a wide number of animal or plant tissues or microorganisms. Three different tube sizes (0.5 mL, 2 mL, or 7 mL) are prefilled with beads for the simple and efficient extraction of DNA, RNA, and protein.1 This automated homogenizer is now being used by a number of groups to release pathogens from their host tissue.
Recently we needed to assess the intestinal load of Caenorhabditis elegans that had been infected with Salmonella typhimurium. We had identified a difference between the survival of two mutants and wild-type worms and wanted to assess whether this was the result of resistance or tolerance of these animals to the infection.2 Previously, another group wishing to examine infection burden had utilized a motorized pestle to homogenize the nematodes.3
We tried a more conventional method—pestle and mortar grinding under liquid nitrogen—which was difficult to control and reasonably hazardous, or disrupting the animals through an 18-guage needle and syringe, but here we found the protective cuticle of the nematode proved difficult to break up. Thus we chose to examine whether the Precellys24 homogenizer could be applied to uniformly disrupt the animals and allow us to quantify the internal infectious load.

Figure 1. Precellys24 homogenizer
Case Studies
Nematodes were infected with a GFP-expressing derivative of S. typhimurium, L1019, which carried a kanamycin-resistance cassette (Figure 2A). At each timepoint, animals were incubated in 200 μL M9 buffer containing 25 mM levamisole hydrochloride (as an anaesthetic; thus preventing pharyngeal pumping and defecation) and 1 mg/mL ampicillin (in order to kill any external bacteria) for 1 h and then washed three times in 200 μL M9 buffer with levamisole hydrochloride.
Animals were disrupted for 10 s in the Precellys24 homogenizer with glass beads (Precellys lysing kit VK05), before the lysates were serially diluted and plated onto LB agar for colonies to be counted by eye (Figure 2B). We were able to conclude that the bacterial burden did not differ between the three animal strains, and that the observed changes in survival were a result of increased tolerance by the mutant nematodes to the infection.2
Others have used the Precellys24 system to examine the viable bacterial load of a Pseudomonas syringae infection of Arabidopsis thaliana,4 and Erwinia carotovora carotovora infection in dissected tissues of Drosophila melanogaster,5,6 respectively. The bacterial growth of virulent and avirulent P. syringae strains was assessed in wild-type and mutant plants lacking a host response factor, PIA1, over four days post-infiltration, to help examine the pathogen genes responsible for the induction of PIA1.
Leaf discs were homogenized in 1 mL sterile water and the subsequent colony forming units (CFUs) were determined by serial dilution.4 In the latter instance, the work had identified the overexpression of antimicrobial peptides in infected Toll-8 (Tollo) mutant larvae, which may have been caused by the overactivation of immune elicitors due to a higher bacterial load in the mutant over control tissues. In order to quantify this burden, CFUs were counted on LB agar, following the disruption of the dissected guts or trachea by 0.75–1 mm glass beads in 500 μL LB broth.6
Moraxella catarrhalis, a Gram negative bacterium that causes otitis media, was shown to elicit an inflammatory response primarily through Toll-like receptor 4 (TLR4) in murine macrophages.7 These experiments were complemented by an in vivo analysis of the role of TLR4 upon M. catarrhalis infection in mice.
Wild-type and TLR4 knockout mice were challenged with M. catarrhalis type A strain 25238, and their lungs were subsequently harvested and homogenized in 1 mL phosphate-buffered saline using the Precellys24. The homogenates were serially diluted and plated onto chocolate agar for bacterial CFU counts, as well as being subjected to an enzyme-linked immunosorbent assay (ELISA) for cytokine detection. The work showed that loss of TLR4 strongly impaired the ability of mice to clear the infection from the lungs.7
The Precellys24 bead beater can also be used to isolate infectious virus from tissue. In order to examine the protective capacity of experimental vaccination for H1N1 in mice, the lungs of immunized animals were removed three days post-viral challenge and homogenized in 1 mL cell media with 2.8 mm ceramic beads (Precellys lysing kit CK28), for 40 seconds, with a 5 second interval between each 20 s, 5,000 rpm burst.
The infective viral titer was subsequently determined by a TCID50 titration on Madin-Darby canine kidney cells.8 The viral titer of the mosquito-borne alphavirus, Chikungunya virus (CHIKV), in infected macaques, has been examined in a parallel manner. 100 mg splenic, hepatic, muscular, or joint tissue was homogenized in the Precellys24 system, again with ceramic beads (Precellys lysing kit CK28) for soft tissues or with metal beads (Precellys lysing kit MK28) for joint and muscle tissues, and the TCID50 assessed using both a mammalian BHK-21 cell line and C6/36 mosquito cells. Together the work suggested that these non-human primates offered a good model for CHIKV pathogenesis.9
In another study, the coxsackievirus B4 (CVB4) titer of infected mice was assessed by qRT-PCR.10 Here the authors were examining the role of matrix metalloproteinases during CVB4-induced pancreatitis.
In this case, the Precellys24 tissue homogenizer was used to prepare tissue lysates from which RNA could be isolated and the viral load quantified. The regime in this instance was the homogenization of 30 mg pancreata tissue in lysis buffer by beating at 6,500 rpm for 3 cycles of 5 seconds, each with intervals of 5 seconds, by ceramic beads (Precellys lysing kit CK28).
In order to ensure the presence of viral RNA originated from infectious virus, rather than a persistent infection, another 30 mg of pancreata tissue was homogenized in saline according to the same procedure; CCID50 was determined by endpoint titration on Vero cell cultures.10
We found the Precellys24 system to be a successful solution to isolating bacteria from the nematode intestine; the main advantages to the technology were the efficiency, speed, and consistency of the homogenization across samples. Of course there will be a need for optimization of the protocol. This brief review already highlights the tissue-dependency of the method both in terms of the beads used and the schedule of homogenization in order to optimally release the pathogen from the surrounding tissue, while maintaining the viability of the microorganism itself.
In conclusion, the Precellys24 tissue homogenizer offers a practical and workable solution to the isolation of intact pathogens from animal or plant tissues.
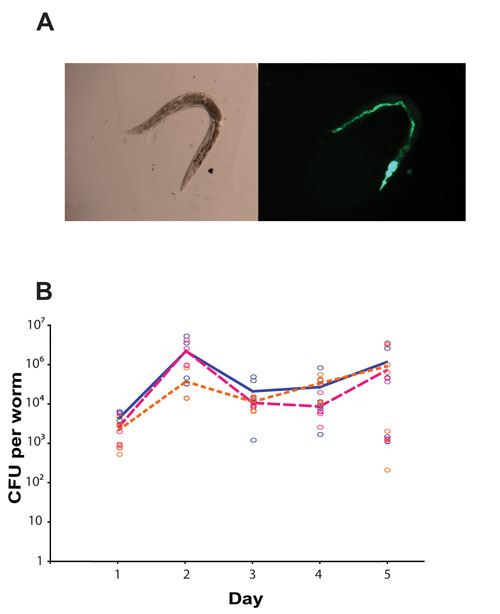
Figure 2. The internal bacterial burden of C. elegans. (A) Bright-field and fluorescence images of a representative animal at day 5 post-S. typhimurium infection (here the bacteria express green fluorescent protein for detection); the pharyngeal structure has been destroyed and the intestine is distended and full of bacteria. (B) The bacterial load per animal was quantified through CFU counts following homogenization with the Precellys24 system. No difference was found between the three animal strains—wild-type animals (blue) and the two mutant strains lacking lys-7 (pink) and abl-1 (orange), each p>0.2.
Elizabeth K. Marsh is post-doctoral fellow at the School of Cancer Sciences, University of Birmingham, Robin C. May is Lister fellow and reader in infectious diseases at the Institute of Microbiology & Infection and School of Biosciences, University of Birmingham, and Esmeralda Carvalho ([email protected]) is application engineer at Bertin Technologies, CNIM Group.
References:
1 R. Verollet, Biotechniques 44 (6), 832 (2008).
2 E. K. Marsh, M. C. W. van den Berg, and R. C. May, PLoS One 6 (3) (2011).
3 T. Kawli and M. W. Tan, Nature Immunology 9 (12), 1415 (2008).
4 I. Widjaja, I. Lassowskat, G. Bethke et al., The Plant Journal 61 (2), 249 (2010).
5 N. Buchon, N. A. Broderick, T. Kuraishi et al., BMC Biology 8, 152 (2010).
6 I. Akhouayri, C. Turc, J. Royet et al., PLoS Pathogens 7 (10) (2011).
7 F. Hassan, D. Ren, W. Zhang et al., PLoS One 7 (5), e37610 (2012).
8 A. Hessel, M. Schewendinger, D. Fritz et al., PLoS One 5 (8) (2010).
9 K. Labadie, T. Larcher, C. Joubert et al., The Journal of Clinical Investigation 120 (3), 894 (2010).
10 A. M. De Palma, E. Verbeken, I. Van Aelst et al., Virology 382 (1), 20 (2008).


