January 1, 2017 (Vol. 37, No. 1)
Novel Technology Allows Platform-Independent Operation
Nucleic acid extraction is the starting point for a large number of processes in molecular biology, making it fundamental to the success of all downstream applications. For many decades, traditional methods—based either on silicate materials (e.g., spin-filter columns), magnetic particle suspensions, or anion exchangers—have been widely used in routine work in research labs.
Now, 35 years after the description of the first silica-based DNA and RNA isolation technique,1 Analytik Jena has introduced SmartExtraction, a novel, next-generation nucleic acid extraction technology. This technology doesn’t only accelerate the entire workflow, it also automates the process to the fullest possible extent. The technique was primarily designed in response to demands for high-quality, high-molecular-weight DNA, an increasingly important material for next-generation sequencing (NGS)2 and other areas as well.
The heart of SmartExtraction is Analytik Jena’s patented DC-Technology® extraction chemistry which offers several benefits, including short lysis times. DC-Technology, which is the basis of all extraction kits available from Analytik Jena, also facilitates the binding of DNA to uniquely modified surfaces for efficient nucleic-acid binding.
Smart modified surfaces, which are introduced as granulates into a 1 mL filter tip currently, can selectively bind the desired nucleic acids and then elute them with a high degree of efficiency. This means that the entire extraction process can take place within the pipette tip. In order to prevent shearing forces effectively, the diameter of the tip opening is several times larger than those traditionally used in liquid handling. In addition, all automation parameters have been selectively optimized for extracting high-molecular-weight DNA.

Figure 1.
High-Molecular-Weight DNA
To study the size distribution of nucleic acids isolated via SmartExtraction, DNA from E. coli was isolated at a concentration of 5 × 106 cells each; after which the nucleic acids were separated via electrophoresis for approximately 25 hours while applying a pulse field (PFGE, Rotaphor, Analytik Jena). For comparison, another sample was processed using an anion exchanger, and the resulting eluates were loaded onto the gel in parallel (Figure 1).
At 284 ng/L, the concentration and quality of the DNA obtained through SmartExtraction is approximately 2.5 times greater than that of nucleic acids extracted with an anion exchanger. This is not the only difference, however: fragment size differs enormously as well. The size of the E. coli DNA obtained via SmartExtraction averages between 48 and 500 kbp, whereas the maximum size of the nucleic acids obtained via anion exchange isolation was only 47 kpb.
The automation capabilities of SmartExtraction can simplify and, in some cases, standardize a large number of workflows. This allows for an uncomplicated extraction process, even on small, automated benchtop instruments such as the GeneTheatre (Analytik Jena) (Figure 2).
The flexible combination of a standard liquid-handling platform and SmartExtraction technology also means that nucleic acids with molecular weights as high as 500 kbp can be isolated with no need for an optimized, robotic extraction system.
SmartExtraction is currently available for isolating from bacteria, yeast, whole blood, and tissue samples/eukaryotic cells, particularly with respect to high-input volume and/or weight. Further kits are in the R&D pipeline and will be launched soon.
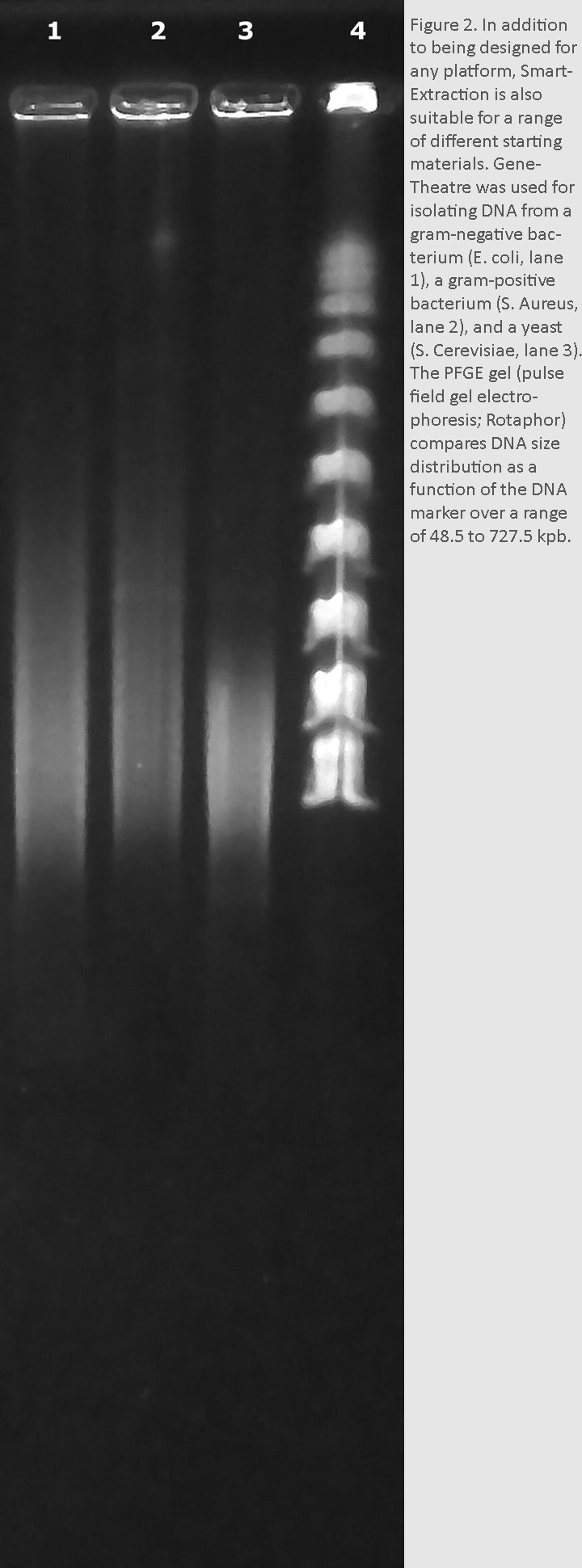
Figure 2.
References
1. Bert Vogelstein, David Gillespie. Preparative and analytical purification of DNA from agarose. Proc. Natl. Acad. Sci. USA. 1979; 76(2): 615–619.
2. Andrea Thürmer. Next Generation Sequencing in der mikrobiellen (Meta)Genomforschung. Biospektrum. 2014; 20: 168–171.
Melanie Kelm serves as head of product management and Matthias Ludwig is an application specialist at Analytik Jena. For more information contact Dana Schmidt ([email protected]).



