October 15, 2017 (Vol. 37, No. 18)
Improve Detection Limits and Preserve Sample Volumes with High-Sensitivity Multiplexing Assays
The future of immuno-oncology treatment and autoimmune diagnostics will involve the detection and quantification of inflammatory biomarkers. As immunotherapy becomes a more common approach for treating cancer and inflammatory disease, cytokines will not only play a role in determining a patient’s disease diagnosis, prognosis, and potential treatment options, but also may serve as a primary index of a therapy’s effectiveness.
Incorporating these biomarkers into clinical practice, however, has been a challenge. Only a few cytokines have been studied in relation to inflammatory disease and immunotherapy, mainly because the biomarkers for these conditions are typically present in very small concentrations, and current research methods aren’t sensitive enough to detect them. It is critical that physicians have the ability to quantify these cytokines to track disease progression before and after treatment.
This tutorial explains how to use the Human Cytokine 6-Plex Panel 1 with the new benchtop Quanterix SR-Plex™ Ultra-Sensitive Biomarker Detection System to simultaneously detect and quantify six inflammatory biomarkers with unprecedented sensitivity using an ELISA-based method. This advanced approach reduces assay volume requirements for multiple sample types and may be sensitive enough to detect circulating disease biomarkers before symptoms appear. As a result, it could potentially provide physicians with a new capability to track disease progression, and the effectiveness of treatments against diseases, which they previously could not monitor with such precision.
Ultra-Sensitive Biomarker Technology
The SR-Plex utilizes Simoa™ (Single Molecule Array) technology, a digital ELISA approach that functions through the same basic mechanisms as traditional ELISAs—but it is 1,000 times more sensitive, which allows it to detect cytokines in the bodily fluids of both healthy and ill individuals.1 The technology derives its sensitivity from dividing the ELISA reaction into tens of thousands of 40-femtoliter-sized wells and measuring the presence of the target molecule separately in each one with single-molecule precision (Figure 1).
In a study from the Journal of Immunological Methods, Simoa technology was proven useful for monitoring therapeutic efficacy—it was the first technique to successfully measure changes in the concentration of circulating Crohn’s disease cytokines following the administration of a specific biological therapeutic.2 Recently, a proof-of-concept study demonstrated applications for this technology in drug-induced liver toxicity. Specifically, researchers were able to rapidly diagnose sepsis in patients and perform pharmacokinetic measurements of interfering nucleic acid therapeutics.3
Digital Method Detects Previously Undetectable Proteins
Simoa technology starts with building an immunocomplex using paramagnetic beads as the solid phase.4 These beads are labeled with fluorescent dyes of various wavelengths and concentrations to create optically distinct subpopulations, and antibodies to specific proteins are immobilized to them. Mixtures of these beads are pooled together to create a multiplex assay.
After they are pooled, the beads are combined with a mixture of biotinylated detection antibodies and added to a 100-µL sample of prediluted sample fluid (typically a 1:4 dilution of blood, urine, or cerebral spinal fluid) in a 96-well plate for a 30-minute incubation. 720,000 beads are added to each sample, resulting in a high bead-to-molecule ratio. The high bead concentration yields two primary advantages:
1. The percentage of beads that contain a labeled immunocomplex follows a Poisson distribution—at low concentrations of protein, the Poisson distribution indicates that each bead will capture either a single immunocomplex or none. For example, if 60,000 protein molecules are captured and labeled on 500,000 beads, then 12% (60,000/500,000) of the beads will carry one protein molecule, and 88% will not carry any protein molecules.
2. With so many beads in solution, the bead-to-bead distance is small, so every molecule encounters a bead in less than a minute. At this time scale, the target analyte molecules, and even large proteins, diffuse quickly. In theory, all the molecules should have multiple collisions with multiple beads.
Next, using an automated plate washer, the magnetic beads are pelleted to remove unbound protein. Beads are then re-suspended in a solution of streptavidin-β-galactosidase (SBG) for 10 minutes and washed twice.
The plate containing the prepped beads is then loaded into the SR-Plex for imaging and analysis. In the SR-Plex, an automated pipetting module uses disposable pipette tips to automatically mix the beads with a solution of resorufin β-d-galactopyranoside (RGP), a fluorogenic substrate of SBG. The mixture is transferred into microfluidic arrays on a Simoa disc, where each sample is loaded into an individual array containing 216,000 femtoliter-sized wells, which have been sized to hold no more than one bead per well (4.25 μm width, 3.25 μm depth). After allowing the beads to settle into the wells, the SR-Plex adds a volume of oil to seal each well of the array, creating a confined volume of 40 fL.

Figure 1. Paramagnetic Simoa beads are used as the solid phase in a sandwich immunoassay containing an enzymatic label. The labeled beads are mixed with a fluorgenic substrate and loaded into an array of femtoliter-size wells on the Simoa disc, where the beads settle into individual wells. The wells are sealed with a layer of oil, which prevents the beads from diffusing out of the wells and allows the fluorescent enzyme product to accumulate at high concentrations, enabling detection of a single enzyme molecule. Fluorescence imaging and data reduction is used to calculate the concentration of each analyte in the assay.
If the target molecule is present (that is, if an immunocomplex has formed), then the captured enzyme label will convert the RGP substrate into a fluorescent product. Since the Simoa disc separates the paramagnetic beads into small volumes, each array contains a high local concentration, enabling the platform to detect accumulated fluorescent RGP products catalyzed by a single enzyme molecule.
The SR-Plex finally captures images of each array on the Simoa disc using LED-based illumination and a 12-bit scientific grade CMOS camera. The camera acquires two fluorescence images of the RGP signal emitted from each array, which enables the SR-Plex software to measure an increase in signal and thereby confirm the presence of a true immunocomplex. It can also distinguish between beads that are associated with a single enzyme molecule (an “on” well) and beads that are not associated with an enzyme (an “off” well). The SR-Plex image analysis software determines the average enzyme per bead (AEB) for both calibrators and samples, and it interpolates the sample concentrations from the calibration curves using its curve-fitting feature.
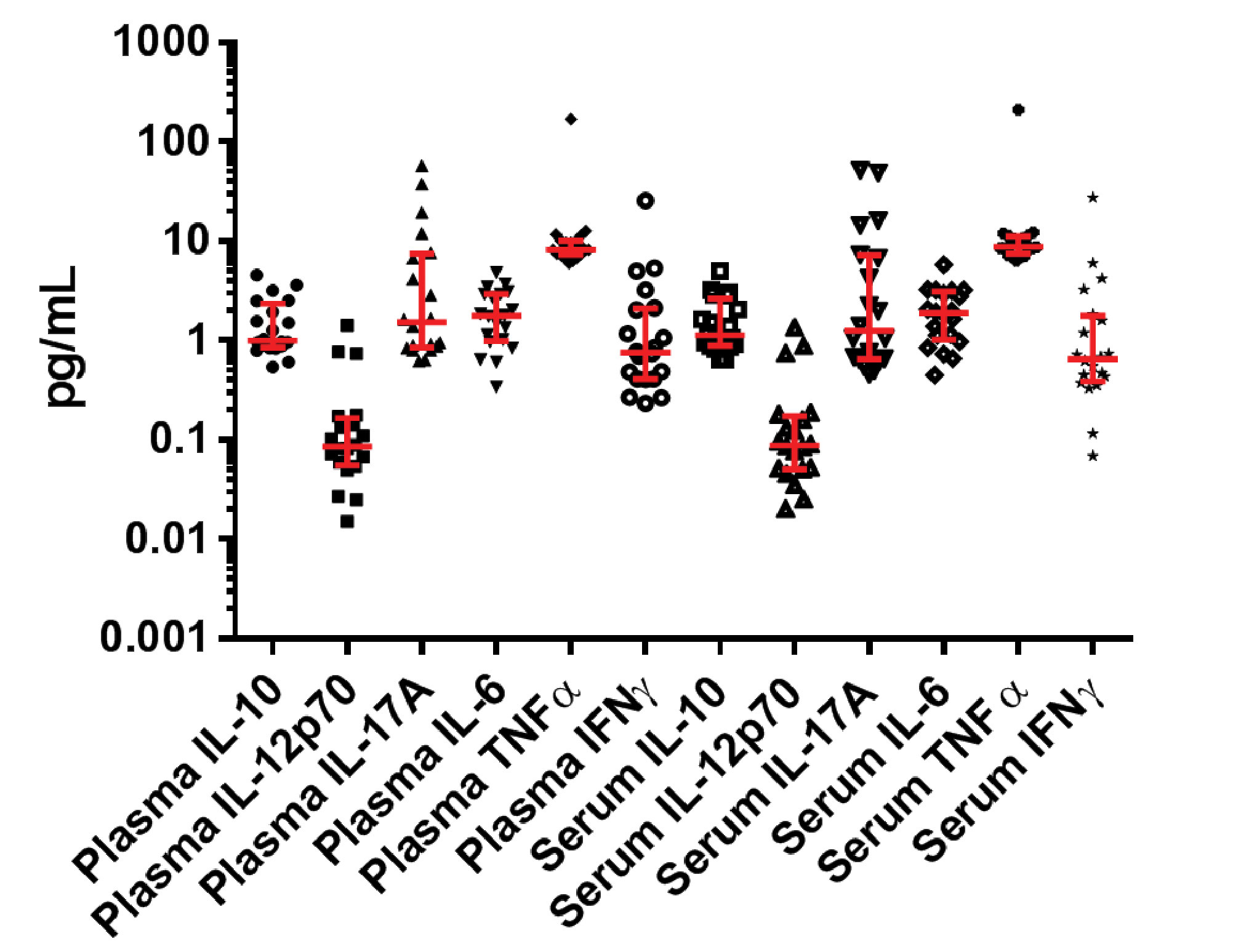
Figure 2. Six inflammatory biomarkers (6 from plasma and 6 from serum for a total of 12 data points) were detected and quantified from 20 healthy donors. Error bars represent median and interquartile ranges.
Improving Inflammatory Biomarker Detection with Multiplexing
Using the SR-Plex, the new HumanCytokine 6-Plex Panel 1 can simultaneously detect and quantify up to six protein biomarkers associated with inflammation and immunotherapy, including biomarkers IL-10, IL-12p70, IL-17A, IL-6, TNFα, and IFNγ, from a single sample. In one case, scientists used this panel to detect and quantify these biomarkers at endogenous levels from healthy donors.5 The assay’s high sensitivity enabled it to detect all six markers in all samples at sub-pg levels (Figure 2). It also exhibited a very low level of cross-reactivity among the biomarkers (Table). The Simoa Human Cytokine 6-Plex Panel 1 is the first commercial immunoassay capable of simultaneously detecting these critical inflammatory biomarkers in serum and plasma at normal levels, providing researchers with a new tool for investigating the role of inflammation in disease progression and the response to immunotherapy.
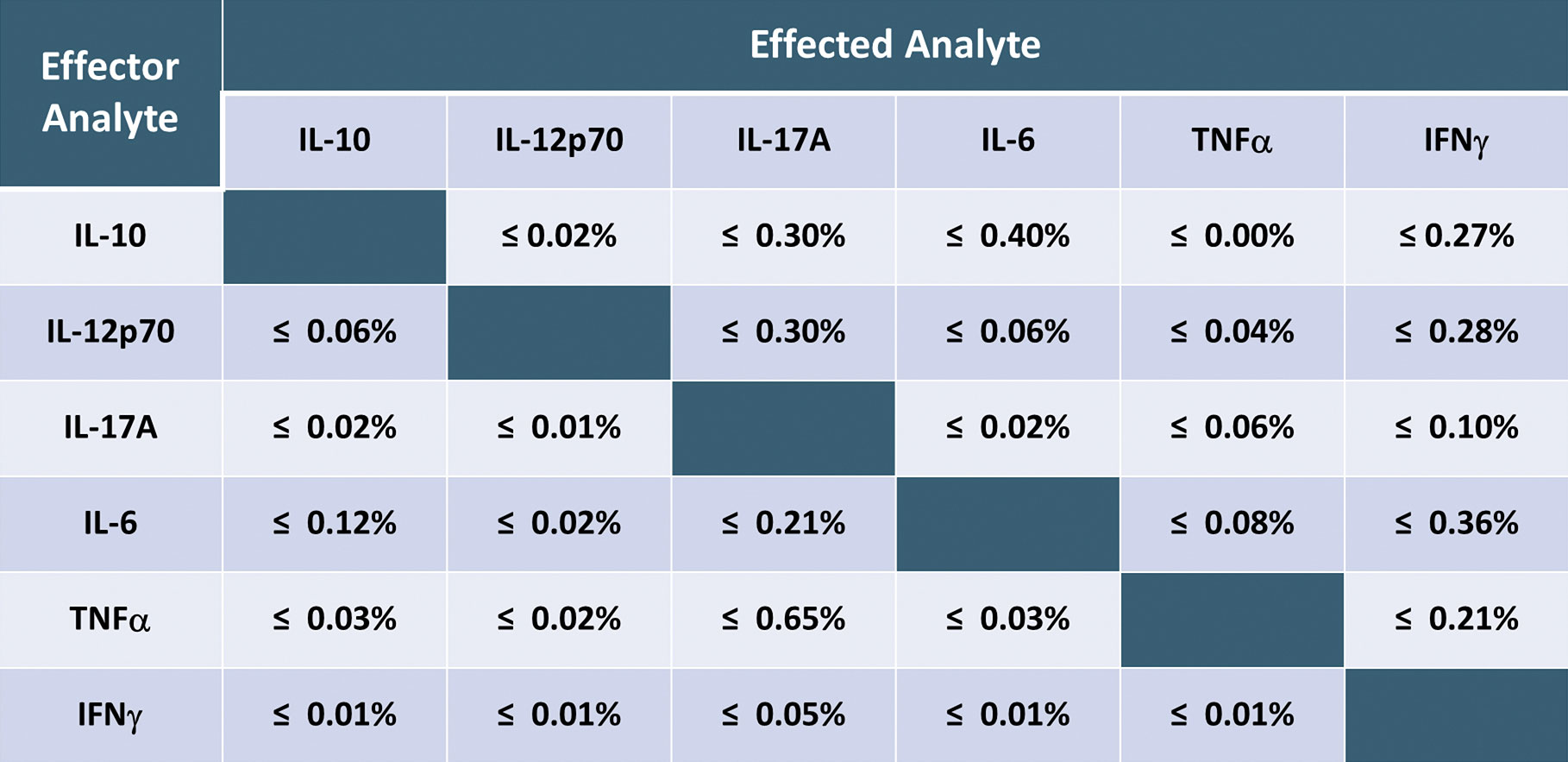
Table. This table shows the high specificity and low level of cross-reactivity between the six biomarkers in the Human Cytokine 6-Plex Panel 1 assay. There was very little cross-talk among the six analytes in the assay.
Conclusions
The SR-Plex with Simoa technology allows researchers to detect and quantify previously undetectable proteins, serum, plasma, CSF, or other sample types. This technology creates the potential to detect biomarkers of disease, such as cancer and inflammatory conditions, much earlier. The newly designed SR-Plex benchtop instrument is an economical option for high-sensitivity biomarker analysis, and may one day yield the discovery of biomarkers with applications across many different fields of clinical research.
1. D. Wu, M.D. Milutinovic, and D.R. Walt, “Single Molecule Array (Simoa) Assay with Optimal Antibody Pairs for Cytokine Detection in Human Serum Samples,” Analyst 140(18), 6277-6282 (2015).
2. Quanterix, “Quanterix Digital ELISA Measures Low Abudance Biomarkers of Inflammation in Crohn’s Disease, accessed on Sept. 1, 2017.
3. D.M. Rissin et al., “Polymerase-Free Measurement of microRNA-122 with Single Base Specificity Using Single Molecule Arrays: Detection of Drug-Induced Liver Injury, PLoS ONE 12(7), e0179669 (2017).
4. Quanterix, “Scientific Principle of Simoa™ (Single Molecule Array) Technology,” White paper, accessed on Sept. 1, 2017.
5. D. Shan et al., “6-Plexed Digital Immunoassay for Immuno-Oncology Markers,” poster presentation, Quanterix; 2017; Lexington, MA, accessed on Sept. 1, 2017.



