The increased diagnosis of central nervous system (CNS) diseases is not matched by increases in therapeutics to treat affected patients. Even for disorders with a highly penetrant genetic component, complex pathogenesis can befuddle drug discovery efforts. For example, scientists have been armed for 25 years with the knowledge that some forms of familial amyotrophic lateral sclerosis (ALS) are linked to superoxide dismutase (SOD1) mutations, but effective therapeutics have not yet been developed. In fact, promising ALS drugs frequently fail in clinical trials, and the few approved drugs prolong life by only a few months. The same disheartening trend is observed for other CNS diseases like autism, Fragile X, Down syndrome, Alzheimer’s, Parkinson’s, and Huntington’s.
Recently, the resurgence of phenotypic screening strategies has revitalized drug discovery. Breakthroughs in cell and molecular biology, robotics, chemistry, and bioinformatics have enabled the use of cells in high-throughput phenotypic screens. Reviews of drug discovery programs conclude that phenotypic screens identify more first-in-class therapeutics as compared to targeted drug discovery. CNS diseases are in desperate need of first-in-class drugs. Further, phenotypic-based drug discovery is continuing to increase in clinical relevance by incorporating disease-specific patient cells.
Utility of iPSC-Derived Neurons in Drug Discovery
Patient-specific cells provide a robustness to high-throughput compound screens that is missing in other disease models, like immortalized cells or mutant cell lines. Even for monogenic diseases, mutant cell lines lack the patient’s genetic background, which influences disease penetrance, onset, and severity, and which, consequently, impact a drug’s therapeutic potential. Historically, the use of patient cells for drug discovery was limited by the amount of material available. However, in 2006, Kazutoshi Takahashi, Ph.D., and Shinya Yamanaka, M.D., Ph.D., discovered how to reprogram somatic cells into “induced” pluripotent stem cells (iPSCs), opening the door for a robust supply of patient-specific cells. Since then, techniques have been developed to amplify patient iPSCs and cleverly differentiate them into almost any cell lineage.
CNS drug discovery is beginning to harness the power of cellular reprogramming to generate large numbers of neurons from patient-derived iPSCs. With extensive libraries of patient iPSCs, diseased neurons can be differentiated for disease-specific screening assays. The onus is now on scientists to design relevant screening assays. Below, we highlight a few important points to consider when using patient iPSC-derived neurons in high-throughput assays.
Technical Considerations and Solutions
An important caveat of using iPSCs in drug discovery is the single origin of the cells. Among iPSC lines there are tremendous variations in transcript profiles, genetic backgrounds, and genetic contributions leading to disease. For practical reasons, usually only one iPSC line is used for initial drug screenings, but other patient lines must be tested in follow-up assays to validate hit compounds. For example, ALS can be sporadic or inherited (familial ALS). Sporadic ALS is “multifactorial,” meaning there is no specific known cause, while familial ALS can have several etiologic mutations in SOD1, C9orf72, TPD43, FUS, or other genes. Hit compounds from screens against a SOD1 patient line must be screened against other ALS patient lines to confirm their broad efficacy.
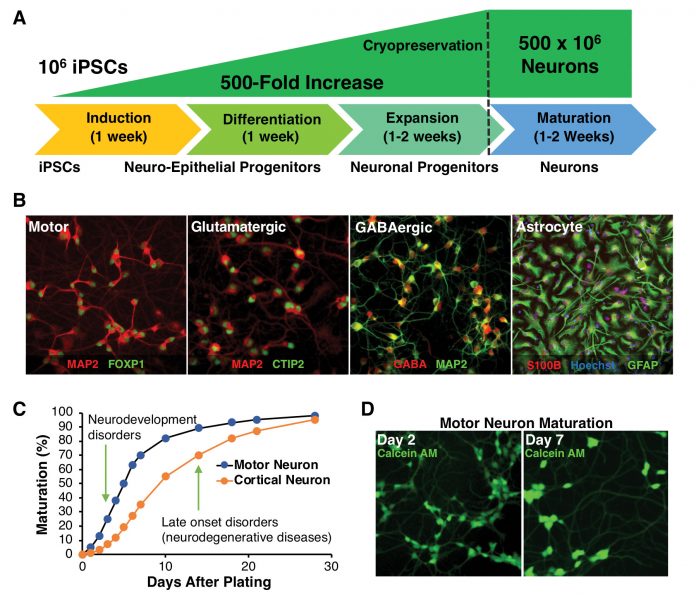
Another important variable in using iPSCs is the integrity and purity of the differentiated neurons. Assays using neurons that are compromised with other cell types or progenitors can muddle phenotypic readouts. For our ALS screening assays, we consistently generate highly pure spinal motor neurons (>90% neuron purity). Similar purity can be achieved with other neuron types including dopaminergic (midbrain), GABAergic (cortical or medium spiny), glutamatergic (cortical or layer V), as well as supportive astrocytes (cortical or spinal) (Figure, A & B). Thus, CNS drug discovery platforms are no longer hampered by previous limits of iPSC differentiation protocols.
Differentiated neurons must be generated consistently and in sufficient quantities for high-throughput screens. In our hands, more than one million motor neurons are used per drug screening plate. The amplification of iPSCs during differentiation regularly produces 500 million motor neurons per batch, and larger yields can be achieved by expanding neuronal progenitors (Figure, A). Critically, each lot of iPSC-derived neurons must meet the same rigorous quality control standards to verify lot-to-lot consistency.
Breakthroughs in gene editing techniques have made isogenic controls possible to help determine disease-specific phenotypes. For example, in familial ALS patient iPSCs, the disease-causing SOD1 mutation at amino acid position 90 (D90A) can be reverted back to the nondiseased sequence (D90D). Thus, the SOD1 D90A diseased motor neuron can be compared to an isogenic D90D control. For other mono- or multigenic diseases, using isogenic controls can help scientists identify and develop phenotypic assays that are specific to the disease.
For high-throughput screens, it is crucial that the plated neurons mature consistently in every well of every plate. Establishing cell-feeding protocols with optimized supplements and neurotrophic factors can improve reliability and predictability of motor neuron maturation. Plate coatings also contribute significantly to neuronal maturation. Coated 384-well plates are commercially available, but coating strategies need to be optimized for 1536-well plates using either amino acid-based (for example, poly-d-Lys) or extracellular matrix proteins (for example, laminin).
Once neuron maturation is defined and consistent, specific assay timepoints can be determined for each CNS disorder and neuron type. Important variables to consider include the emergence of disease phenotype, timing of compound delivery, and length of treatment before measuring the extent of phenotypic rescue. Neurons from patients with early-onset CNS diseases may exhibit phenotypes quickly after plating, whereas late-onset neurodegenerative phenotypes may necessitate prolonged maturation (Figure, C & D).
Screening compounds against neurodegenerative diseases may require one to three weeks of neuron maturation and an additional week of drug treatment. Prolonged incubation times complicate high-throughput screening platforms because they increase evaporation-dependent edge effects and the number of user interventions. Evaporation can be mitigated by incubating 384- or 1536-well plates within a secondary humidified box inside a humidified incubator or developing an active humidification system that uses mist to increase incubator humidity. Prolonged maintenance of neurons requires medium changes and can make high-throughput assays logistically impractical. For example, we found that ALS-specific phenotypes emerged within a few days after neuron plating. So, even though assay performance was best after a week of neuron maturation, we were able to streamline the protocol to four days and avoid medium changes for screening large libraries of compounds.
The final drug screening assay consideration we broadly discuss is the choice of phenotype and reporter system. CNS diseases have phenotypes and pathogeneses that are still debated, so we cannot recommend specific strategies. However, it is important to think critically about which phenotype appears causal to disease onset or propagation and then develop highly responsive assays to detect those changes. Fortunately, scientists have a plethora of options for assay development, as new technologies are constantly emerging for luminescence, fluorescence, and other high-throughput-compatible detection modalities.
Conclusion
The ability to differentiate large numbers of neurons consistently and predictably from patient-derived iPSCs has improved the clinical relevance of drug screening paradigms. With careful selection of assay set-up and controls, the only remaining limit is the scientists’ ability to identify and design disease-specific assays. We are excited to see improvements in drug screening design that will produce greatly needed therapeutics for a variety of challenging and currently untreated CNS disorders.
Paul Guyett, Ph.D., is application scientist, Mike Hendrickson is project manager, and Kurt Laha, Ph.D. ([email protected]), is product manager at BrainXell.

