June 15, 2007 (Vol. 27, No. 12)
Processing Hundreds of Microplate Equivalents Without Complex Plate-Handling Equipment
The requirement for increased throughput in SNP genotyping has led to the development of several technologies that do not use microtiter plates, such as gene chips and bead-based systems. These methods offer extremely high throughput but often with the sacrifice of the flexibility available with plate-based assays. The use of array tape in place of microplates offers a miniaturized, high-throughput automated format for SNP genotyping that maintains the flexibility of plate-based methods.
Array tape (Figure 1) is a continuous plastic tape in which standard SBS format 96-, 384-, or 1,536-well arrays are serially embossed. The tape is used in conjunction with a flexible configuration of dispensing, pipetting, sealing, and detection modules (Figure 2), manufactured by Douglas Global Array (www.global-array.com). This allows for automated, continuous processing of hundreds of microplate equivalents without the requirement for robots or complex plate-handling equipment.
Array tape is a variation of a product called carrier tape, which is used to package integrated circuits in the electronics industry. Prior to the 1980s, integrated circuits and other components were packaged in individual trays or tubes. With the introduction of carrier tape, components were stored and transported in wells on reels of tape, held in place by a continuous cover tape. This greatly simplified the automation of printed circuit board manufacturing. The use of carrier tape is currently the electronics industry standard.
Array tape technology was developed and first implemented in the field of genetic testing by James Weber, Ph.D., previously of the Marshfield Clinic and president of PreventionGenetics (www.preventiongenetics.com). Dr. Weber’s lab at the Marshfield Clinic successfully prototyped and manufactured the first working models of array-tape-based dispensing, pipetting, sealing, and detection equipment.

Figure 1
Cost-savings vs. Microplates
In addition to throughput capacity, a main advantage of array tape over microplates is cost savings on consumables, reagents, and labor (Figure 3). The tape and seal cost less than half the price of corresponding microplates and lids. Reaction volumes in tape are typically between 10–20% of those in microplates, leading to an 80–90% cost savings on reagents. Also, the number of plate equivalents a given technician can process per shift using array tape is about five times the number he or she can process using microplates.
Array tape is especially well-suited for PCR-based processes. Thermal cycling is performed on sealed array tape in a sequential dipping-style water bath thermal cycler. This allows for simultaneous processing of hundreds of microplate equivalents at one time, and provides rapid temperature cycling in comparison with plate-based methods. The array tape is made of a 0.3-mm-thick polypropylene, and the heat transfer through this material is much faster than through typical microplates. During each cycle, samples reach the ambient water bath temperature more quickly and spend a greater percentage of time at the goal thermal-cycling temperature. This allows for shorter cycle times and should aid in reducing nonspecific binding.
The flat contour of the top surface of the tape allows for secure fixation of pressure-sensitive sealing tape over the entire array. A bottom roller mechanism with indentations for each well allows the top roller to apply pressure between the wells as well as around the perimeter of the 384-well footprint. This creates a secure seal with a leakage rate in the water bath thermal cycler that is essentially zero.
The method by which array tape is manufactured (embossing rather than injection molding) makes it relatively inexpensive to customize well size, shape, and configuration (Table).

Figure 2
SNP Genotyping on Array Tape
PreventionGenetics has converted all of its SNP and insertion-deletion polymorphism genotyping, previously performed in low-volume, 384-well PCR microplates, to processes performed in Global Array 384-well polypropylene 1.5-µL array tape. This has involved reducing the reaction volume from 5 µL to about 800 nL. It was found that for both allele-specific and TaqMan assays, using the same concentrations of reagents as in plates in lower volumes in tape yields good results. While reaction volumes of as little as 300 nL have yielded scorable genotypes, an 800-nL reaction volume yields more consistent results. The final reaction protocol for allele-specific PCR assays in array tape is as follows: 5 ng of template DNA in 800nL of 10mM Tris, 0.2mM EDTA, pH of 8.0 is pipetted into the 384 well array tape and dried. 800 nL of a common reagent solution containing 1 X PCR buffer, 600 nM of common primer, 45 nM each of allele-specific primer, 2.5mM Mg+2, 37.5 nM Fam UniPrimer, 50 nM Joe UniPrimer (Chemicon), 250 µM dNTPs, 1% Rox reference dye solution (Invitrogen), and 0.05 units/µL Platinum Taq® polymerase (Invitrogen) is then added to the entire array. A sealing cover tape is applied. The samples are amplified in a water bath thermal cycler under the following conditions: 95°C x 2 minutes, followed by 35 cycles of 95°C x 20 seconds, 55°C x 40 seconds, 72°C x 20 seconds, followed by a final extension at 72°C x 6 minutes.
The TaqMan protocol is basically similar to the above allele-specific PCR protocol, with the following variations: 5 ng of template DNA is pipetted into the plate and dried. 800 nL of a reaction mixture containing 1 X PCR buffer, 100 µM dNTPs, 0.05 units/µL Platinum Taq®, 1.5 mM Mg+2, 1% Rox reference dye solution (Invitrogen), and 0.15 X TaqMan primers and probes (primers: 0.134 µM, probes: 0.030 µM) is then added to the dried DNA in the tape. The samples are amplified in a water bath thermal cycler with the following cycle pattern: 95°C for 3 minutes, then 35 cycles of 95°C x 20 seconds, 60°C x 1 minute, with a final incubation at 60°C for 6 minutes.
The prethermal cycling processes of pipetting and drying template DNA, dispensing reaction mixture, and applying a sealing cover tape are performed in a continuous process on sequential Douglas Scientific modules. The template DNA is pipetted from sample plates into tape using a 384-tip syringe type pipettor (Roche).
The samples are then passed over drying modules where all liquid is evaporated from the DNA samples. A Douglas Scientific jet-type dispenser delivers the reaction mixture. A low autofluorescence polyolefin clear Global Array cover tape is then applied. The sealed arrays are transferred to a water bath thermal cycler for amplification. Following thermal cycling, arrays are transferred to a detection module, and detection is performed through the cover tape.
PreventionGenetics has developed its own fluorescence readers that feed tape in a continuous fashion. The raw data are processed and scored with software developed in-house. A commercial detection unit is available through Douglas Scientific.
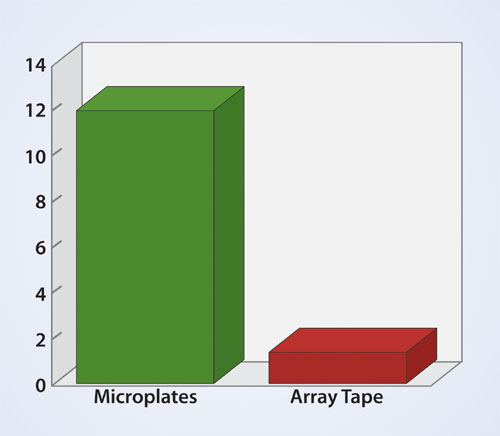
Figure 3
Discussion
Array tape automation provides an efficient, miniaturized format for high-throughput genotyping. It is an alternative to chips and bead-based systems for use in applications where these methods are not convenient or practical. Because of the capacity for high-throughput thermal cycling, it is ideal for situations such as fine mapping where many thousands of samples are tested on a few hundred or less SNPs.
Elaine May, M.D., is CSO at Global Array. Web: www.global-array.com. Phone: (952) 886-0987.
E-mail: [email protected].



