Scientists have developed virtual reality software that allows researchers to walk around and analyze the interiors of individual cells. The software, called vLUME—visualization of the local universe in a microenvironment and pronounced “volume”—allows super-resolution microscopy data to be visualized and analyzed in virtual reality, and can be used to study everything from individual proteins to entire cells. Created by scientists at the University of Cambridge in the U.K., and at 3D image analysis software company, Lume VR, the platform could help researchers understand fundamental problems in biology, and develop new therapeutic strategies. Th vLUME software is being made available free for academic use.
“Biology occurs in 3D, but up until now it has been difficult to interact with the data on a 2D computer screen in an intuitive and immersive way,” said research lead Steven F. Lee PhD, at the University of Cambridge, department of chemistry. “It wasn’t until we started seeing our data in virtual reality that everything clicked into place.” Lee and colleagues at Lume VR, the University of Leeds, the University of Oxford, Aix Marseille Université, Instituto Gulbenkian de Ciência, and University College, London, reported on the vLUME platform in Nature Methods, in a paper titled “vLUME: 3D virtual reality for single-molecule localization microscopy.”
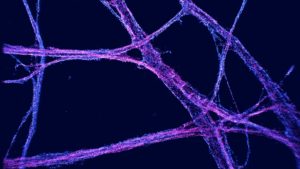
However, while the technology has allowed researchers to observe molecular processes as they happen, one problem has been a lack of ways to visualize and analyze the data in three dimensions. “ … there is now a marked absence of 3D visualization approaches that enable the straightforward, high-fidelity exploration of this type of data,” the team continued.
The vLUME project started when Lee and his group met with the Lume VR founders at an event at the Science Museum in London. While Lee’s group had expertise in super-resolution microscopy, the team from Lume specialized in spatial computing and data analysis. By combining their respective expertise, the researchers have developed vLUME into a powerful new tool for exploring complex datasets in virtual reality. They describe the platform as “…an immersive virtual reality (VR)-based visualization software package purposefully designed to render large 3D-SMLM datasets.”

“vLUME is revolutionary imaging software that brings humans into the nanoscale,” said Alexandre Kitching, CEO of Lume. “It allows scientists to visualize, question, and interact with 3D biological data in real-time, all within a virtual reality environment, to find answers to biological questions faster. It’s a new tool for new discoveries.”
Viewing data in this way can stimulate new initiatives and ideas. Anoushka Handa, a PhD student in Lee’s group, used the software to image an immune cell taken from her own blood, and then stood inside her own cell in virtual reality. “It’s incredible—it gives you an entirely different perspective on your work,” she said.
The software allows multiple datasets with millions of data points to be loaded in and finds patterns in the complex data using in-built clustering algorithms. The findings can then be shared with collaborators worldwide using image and video features in the software. “Data generated from super-resolution microscopy is extremely complex,” said Kitching. “For scientists, running analysis on this data can be very time consuming. With vLUME, we have managed to vastly reduce that wait time allowing for more rapid testing and analysis.”
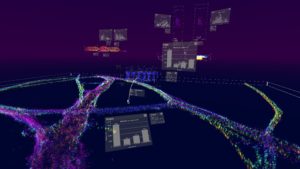
The software can be used by people with little prior expertise. “It enables imaging scientists with any level of expertise to make straightforward analytical sense of what is often highly complex 3D data,” the team noted. “We envisage that users will use vLUME to share and explore existing and new emergent datasets and that the ability to interact with SMLM data in this way may inspire new methods, tools, and discoveries. Future directions could include the incorporation of a multi-user tool for numerous users to use vLUME within the same environment, as well as the incorporation of advanced computation imaging tools such as focused training methods for machine learning.”



