Sometimes a replicating cell must choose the lesser of two evils. For example, when a replicating cell is stressed, it may struggle to pair bases against its single-stranded DNA (ssDNA) template, even if that template is flawless. Faced with the risk of leaving a DNA strand uncopied and open to catastrophic DNA rearrangements, the cell may resort to a handy expedient. Rather than wait for highly accurate DNA polymerase enzymes, the cell may call on a hastier but error-prone substitute—mutagenic repair.
Yes, mutagenic repair, which relies on translesion synthesis (TLS) DNA polymerases, may intrude even when DNA is free of mutations. This finding comes from the University of Toronto, where scientists noticed that in times of stress, yeast cells produce far more replication errors than would have been expected. These errors, which occurred because the yeast cells were deprived adequate supplies of nucleotide bases, could also occur in our own cells, leading to cancer and other diseases.
The University of Toronto scientists, led by Grant W. Brown, PhD, suggested that mutagenic repair and its error-prone polymerases could be replicating more of our DNA than we ever suspected, and that these polymerases could be the source of disease-causing mutations.
Details of the scientists’ work appeared February 4 in the journal Molecular Cell, in an article titled, “Rad5 Recruits Error-Prone DNA Polymerases for Mutagenic Repair of ssDNA Gaps on Undamaged Templates.” The article notes that ordinarily, TLS is activated in post-replication repair (PRR) when physical damage is detected. Such damage may be caused by UV light or carcinogenic chemicals. Yet PPR may also be activated to repair ssDNA resulting from lesion-less replication stress.
“We find that PRR responds to DNA replication stress that does not cause base lesions,” the article’s authors wrote. “Rad5 forms nuclear foci during normal S phase and after exposure to types of replication stress where DNA base lesions are likely absent. Rad5 binds to the sites of stressed DNA replication forks, where it recruits TLS polymerases to repair ssDNA gaps, preventing mitotic defects and chromosome breaks.”
The discovery took Brown and David Gallo, a PhD student who did most of the work, by surprise. They were studying DNA replication in cells raised under a limited supply of nucleotide bases that make up the A, T, C, and G letters of the DNA code. “You can think of it as your car running out of fuel. We were removing gas from DNA replication,” explained Gallo.
Cells may encounter this type of stress when food is scarce or in disease, when resources are exhausted by fast-proliferating cancer cells, for example.
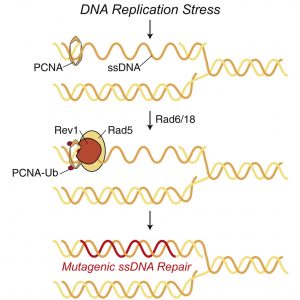
To make a copy of their genome, cells first unwind the double DNA helix into single strands, where each strand serves as a template against which new DNA is synthesized through complementary base pairing, A with T and C with G. This is usually done by DNA polymerase enzymes which are “super accurate and only make errors very rarely to ensure the blueprint of life is passed to the next generation with high fidelity,” Brown noted.
But the lack of DNA building blocks led the cells to call on another type of DNA polymerase, which is more fallible. This expedient, mutagenic repair, may seem an odd way to preserve the DNA, but it helps avoid worse genome-wrecking scenarios in which unwound DNA strands can lead to portions of chromosomes being lost.
“It’s better to copy and make some mistakes than to leave it uncopied and open to chromosomal rearrangements which would be much worse for the cell,” Brown emphasized.
“In contrast to the prevailing view of PRR,” the authors of the Molecular Cell article concluded, “our data indicate that Rad5 promotes both mutagenic and error-free repair of undamaged ssDNA that arises during physiological and exogenous replication stress.” This finding could have implications for cancer research.
Our cells have the same error-prone DNA copying machinery. And fast-proliferating cancer cells often experience what’s known as oncogene-induced replication stress, when they run out of fuel as DNA replication rates outstrip nucleotide supply. Under these conditions, the cells could resort to error-prone DNA replication, where new mutations could help cancer survive although this remains to be verified by future studies.
“The error-prone DNA replication pathway could very well be activated during oncogene-induced replication stress to help cancer cells survive,” Gallo pointed out. “This would make it a hot therapeutic target to selectively kill cancer cells.”




