Individuals with Usher syndrome (USH) are born deaf and begin to suffer retinal degeneration from puberty, eventually losing their eyesight as the disease progresses. USH is the most common cause of combined hereditary deafness and blindness affecting one in every 6,000 people worldwide.
For the past 25 years, a research group led by Uwe Wolfrum, PhD, professor at the Institute of Molecular Physiology at Johannes Gutenberg University Mainz (JGU) has been investigating the mechanisms underlying Usher syndrome.
In collaboration with a group led by Reinhard Lührmann, PhD, professor at the Max Planck Institute for Biophysical Chemistry in Göttingen, Wolfrum’s team has now identified a novel pathological mechanism leading to USH.
The team has discovered that the protein SANS synthesized from a gene called Usher syndrome type 1G (USH1G), plays a crucial role in regulating the common cellular process called splicing that connects different coding sequences of a gene to produce multiple proteins. The researchers show defects in the SANS protein can lead to errors in the splicing of genes related to cell division and USH.
The study is published in an article in the journal Nucleic Acids Research titled, “SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes.” Financial support for the study came from GeneRED, IPP Programme Mainz, FAUN-Stiftung, Nuremberg, Foundation Fighting Blindness (FFB), and RP Fighting Blindness.
“We are aiming to elucidate the molecular basis that leads to the degeneration of the light-sensitive photoreceptor cells in the eye in cases of Usher syndrome,” said Wolfrum.
Cochlear implants can compensate for hearing loss in patients with USH but there are currently no treatments for USH-associated blindness.
Earlier studies from Wolfrum’s team have shown that SANS acts as a scaffold protein with multiple domains on which other proteins dock, ensuring proper cellular function. SANS mutations can lead to malfunctions of the auditory and vestibular hair cells in the inner ear and of the photoreceptor cells of the retina responsible for the sensory defects in USH.
Until now, how SANS contributes to pathogenic processes in the eye had remained unclear. SANS is expressed in the photoreceptors of the retina and supporting glia cells.
“So far, we had thought of SANS simply as a scaffold molecule that participates in transport processes in the cytoplasm associated with ciliary extensions,” said Wolfrum. “But recently, Adem Yildirim in his PhD thesis conducted in the International PhD Program (IPP) in Mainz discovered that SANS interacts with splicing factors to regulate pre-mRNA splicing.”
During splicing, non-coding sections of a gene (introns) are removed from transcribed pre-mRNA and in alternative splicing, coding sections of a gene (exons) that are not required for a specific protein variant are excluded as well. The resulting messenger RNA is then used for protein biosynthesis.
The splicing process is catalyzed in the nucleus by the spliceosome, a dynamic, highly complex molecular machine that is assembled during the splicing process from a number of subcomplexes of protein and RNA components.
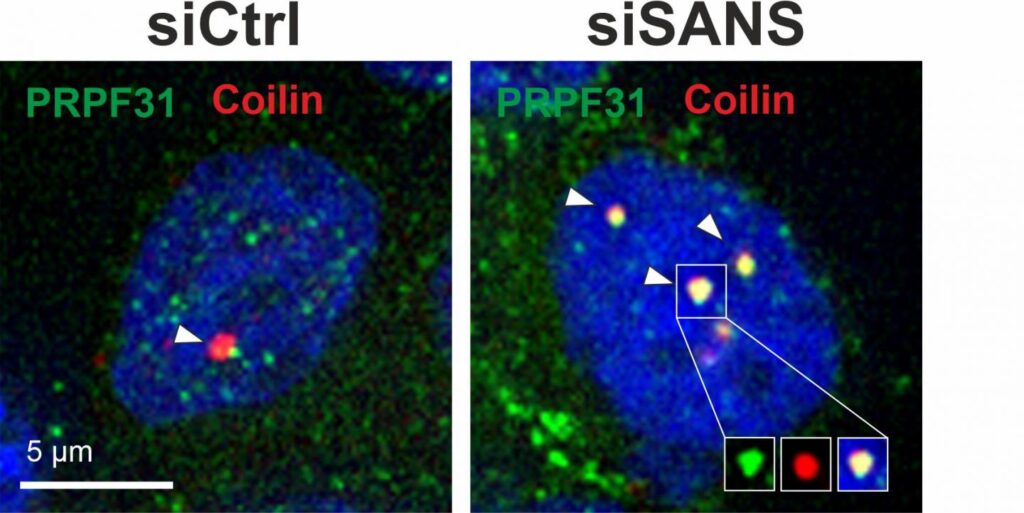
The current study fount that in the nucleus SANS is responsible for transferring components of spliceosome subcomplexes (tri-snRNP complexes) from the Cajal bodies (spherical bodies of RNA and protein) to nuclear speckles (dynamic subnuclear structures containing pre-messenger RNA splicing factors). In the nuclear speckles, tri-snRNP complexes bind to the spliceosome assembly to subsequently activate it.
Lack or dysfunction of SANS prevents the spliceosome from being correctly assembled and activated which leads to incorrect splicing of other Usher syndrome-related genes, ultimately leading to the clinical manifestation of the disorder.
The authors noted, “We provide the first evidence that splicing dysregulation may participate in the pathophysiology of Usher syndrome.”
“In addition to the new findings relating to the splicing mechanism, we have also identified new aspects that we aim to investigate with regard to developing concepts for the treatment and therapy of the Usher syndrome in future,” said Wolfrum.