
If you’ve ever spent time in a molecular biology lab, there’s a good chance you’ve opened up a fridge box filled with restriction enzymes. These days, however, may soon be a thing of the past, as CRISPR–Cas nucleases cleave DNA in a programmable manner, offering advantages over restriction enzymes for in vitro molecular applications.
But CRISPR-Cas nucleases also have their pitfalls, notably as the requirement to recognize a short DNA motif adjacent to the target site, known as protospacer adjacent motifs (PAMs). The dependence on the availability of a PAM proximal to the target site prohibits precise cleavage and limits the sequence targetability of DNA substrates.
A new study demonstrates that a SpCas9 variant, SpRY, is PAMless in vitro and can cleave DNA at practically any sequence, including sites refractory to cleavage with wild-type SpCas9. Led by Kathleen A. Christie, PhD., a postdoctoral researcher at Harvard Medical School and research fellow at Massachusetts General Hospital, the study illustrates the versatility and effectiveness of SpRY DNA digests (SpRYgests) to improve the precision of several cloning workflows, including those not possible with restriction enzymes or canonical CRISPR nucleases.
The article “Precise DNA cleavage using CRISPR-SpRYgests” was published in the October issue of Nature Biotechnology.
I don’t want any PAM!

The nuances and challenges of molecular cloning techniques have been a battle for many researchers, including senior author Ben Kleinstiver, PhD, assistant investigator at Mass General Hospital and Assistant Professor at Harvard Medical School. “Being beholden to a catalog of restriction enzymes (that even collectively don’t permit cutting DNA anywhere you want) was frustrating,” Kleinstiver told GEN. “During my postdoc in Keith Joung’s lab, an intern, Nicolas Wyvekens, and I dreamed about ways to try to repurpose argonaute proteins to enable simpler and unconstrained DNA digests.”
Although other groups have made progress in that area, several challenges remain. When Kleinstiver started his lab a few years ago, the team sought to build the first CRISPR-Cas enzyme that could read the entire genome and no longer require a specific protospacer adjacent motif (PAM) to edit DNA. In a Science article published in 2020, the group demonstrated the development of SpRY. In this work, Kleinstiver’s team found that in human cells, SpRY preferred sites with NRN PAMs and had a weaker ability to target sites with NYN PAMs (where N = any base, R = A or G, and Y = C or T).
“From there, it dawned on us that we could use this enzyme for many things other than DNA editing in human cells,” said Kleinstiver. “So, we turned our attention to repurposing the PAMless SpRY for molecular cloning applications since SpRY’s lack of sequence requirements outside of its gRNA should, in principle, overcome the incomplete retargetability and other issues of prior approaches (like restriction enzymes, argonaute proteins, or conventional CRISPR-Cas enzymes).”
The humble SpRY
When testing SpRY in vitro, the research team, led by Christie, was surprised to find it was essentially PAMless. This discovery overcame the remaining hurdle of cutting DNA at any specified base.
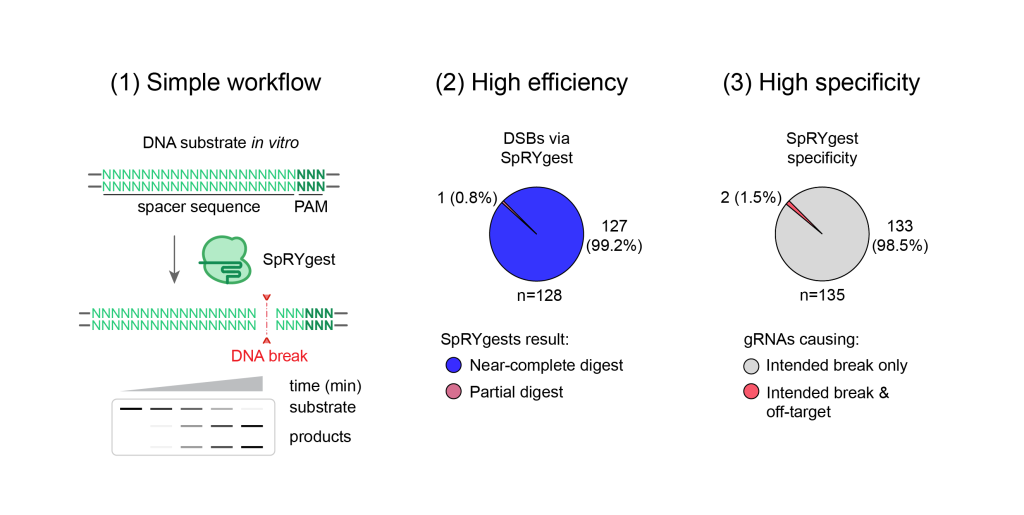
The method will be published in our manuscript, and Kleinstiver hopes it will allow anyone to practice SpRYgests. “The oligonucleotides can be purchased from vendors, the gRNA transcription kits are already commercially available, and we hope that soon the protein will be available from vendors as well (but in the meantime, we have provided details to purify the SpRY enzyme),” said Kleinstiver. “We hope that the simplicity of this method will permit users to incorporate SpRYgests into their typical molecular cloning workflows without too many challenges.”
SpRYgest: The best in vitro DNA digest?
Kleinstiver envisions a streamlined SpRY kit where users could simply plug an oligo into a solution would also be convenient, enabling SpRYgests to be implemented in various applications. “For example, for typical methods like molecular cloning where segments of plasmids are interchanged, SpRYgests can be paired with isothermal assembly methods (i.e., Gibson Assembly) to simplify and improve the precision of the approach,” Kleinstiver told GEN.
Kleinstiver said that SpRYgest would also simplify and expedite more complex cloning methods like constructing saturation mutagenesis libraries. “To have these libraries synthesized by companies can be very expensive (in the thousands to tens of thousands of dollars),” Kleinstiver told GEN. “We show that SpRYgest enables the generation of these libraries in a matter of days for a few dozen dollars.”
Beyond molecular cloning, Kleinstiver said that SpRYgests unlock the ability to cut DNA at any user-specified position, which can be useful for DNA library construction or sequence depletion (for next-generation sequencing approaches) and many other applications.



