Neurological drugs that have difficulty crossing the blood-brain barrier might fit into the hold of a major facilitator superfamily domain containing 2A (MFSD2A)—a sort of molecular ferry. Ordinarily, MFSD2A transports omega-3 fatty acids into the brain. But now that scientists based at Columbia University have determined MFSD2A’s three-dimensional structure, they are hopeful that drugs could be designed that would be compatible with the transporter’s specifications. For such drugs, the impassable would become, well, passable.
The three-dimensional structure, which was determined using single-particle cryo-electron microscopy, shows how omega-3s bind to the transporter. “This information may allow for the design of drugs that mimic omega-3s to hijack this system and get into the brain,” said Rosemary J. Cater, PhD, a research fellow in the Columbia University laboratory led by Filippo Mancia, PhD, an associate professor of physiology and cellular biophysics.
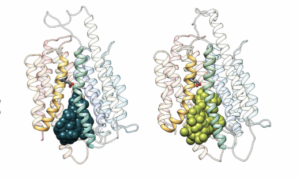
Cater is the first author and Mancia is the senior author of a study (“Structural basis of omega-3 fatty acid transport across the blood–brain barrier”) that appeared June 16 in the journal Nature. According to this study, MFSD2A has 12 transmembrane helices that are separated into two pseudosymmetric domains.
“The transporter is in an inward-facing conformation and features a large amphipathic cavity that contains the Na+-binding site and a bound lysolipid substrate, which we confirmed using native mass spectrometry,” the article’s authors wrote. “Together with our functional analyses and molecular dynamics simulations, this structure reveals details of how MFSD2A interacts with substrates and how Na+-dependent conformational changes allow for the release of these substrates into the membrane through a lateral gate.”
These results, the authors suggest, could “provide a foundation for the structure-based design of neurotherapeutic agents that hijack MFSD2A for delivery across the blood-brain barrier, which is currently a major bottleneck in the development of neurotherapeutic drugs.”
The blood-brain barrier is a layer of tightly packed cells that lines the brain’s blood vessels and zealously blocks toxins, pathogens, and some nutrients from entering the brain. Unfortunately, the layer also blocks many drugs that are otherwise promising candidates to treat neurological disorders.
Essential nutrients like omega-3s require the assistance of dedicated transporter proteins such as MFSD2A. “The transporters are like bouncers at a club, only letting molecules with invites or backstage passes in,” Cater explained. “Understanding what MFSD2A looks like and how it pulls omega-3s across the blood-brain barrier may provide us with the information we need to design drugs that can trick this bouncer and gain entry passes.”
To visualize MFSD2A, Cater used a technique called single-particle cryo-electron microscopy (cryo-EM).
“The beauty of this technique is that we’re able to see the shape of the transporter with details down to a fraction of a billionth of a meter,” said Mancia. “This information is critical for understanding how the transporter works at a molecular level.”
For cryo-EM analysis, protein molecules are suspended in a thin layer of ice under an electron microscope. Powerful cameras take millions of pictures of the proteins from countless angles which can then be pieced together to construct a 3D map.
Into this map, researchers can build a 3D model of the protein, putting each atom in its place. “It reminds me of solving a jigsaw puzzle,” Mancia remarked. This technique has become remarkably powerful in visualizing biological molecules in recent years, thanks in part to Joachim Frank, PhD, professor of biochemistry and molecular biophysics at Columbia University Vagelos College of Physicians and Surgeons, who won the Nobel Prize in 2017 for his role in developing cryo-EM data analysis algorithms.
“Our structure shows that MFSD2A has a bowl-like shape and that omega-3s bind to a specific side of this bowl,” Cater explained. “The bowl is upside down and faces the inside of the cell, but this is just a single 3D snapshot of the protein, which in real life has to move to transport the omega-3s. To understand exactly how it works, we need either multiple different snapshots or, better yet, a movie of the transporter in motion.”
A third co-leader of the study, David Silver, PhD, professor at the Duke-NUS Medical School in Singapore and pioneer in MFSD2A biology, together with his team tested and confirmed hypotheses derived from the structure and the computational simulations on how MFSD2A works to pinpoint specific parts of the protein that are important.
The team also included researchers from the New York Structural Biology Center, the University of Chicago, and the University of Arizona, all using their specific skills to make this project possible.
The team is now investigating how the transporter first recognizes omega-3s from the bloodstream. “But our study has already given us tremendous insight into how MFSD2A delivers omega-3s to the brain,” Cater noted. “We are really excited to see where our results lead.”


