Scientists at Uppsala University say they have developed a new technique to determine how effective two antibiotics combined can be in stopping bacterial growth. The new method is simple for laboratories to use and can provide greater scope for customizing treatment of bacterial infections, according to the researchers whose study “CombiANT: Antibiotic interaction testing made easy” appears in PLOS Biology.
Combinations of antimicrobial agents are invariably prescribed for certain infectious diseases, such as tuberculosis, HIV, and malaria. Bacterial infections that are not readily treatable, such as those affecting cardiac valves and prostheses, and lung infections in cystic fibrosis, are also usually subjected to a combination of antibiotics.
Bottom line is that synergism is more effective than could in fact have been expected, based on the efficacy of the separate agents. In contrast, the opposite phenomenon, i.e., two antibiotics counteracting each other’s effects is undesirable. However, knowing what the combined effect will be is not always easy.
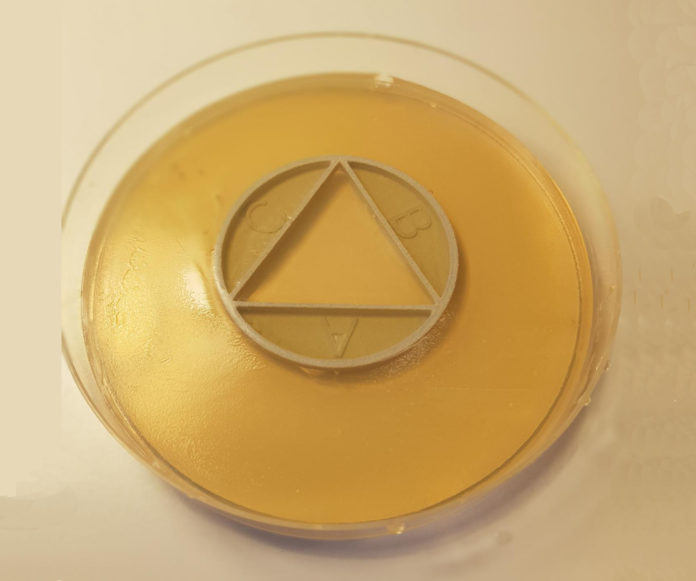
With the newly developed method known as CombiANT (combinations of antibiotics), interactions between various antibiotics can be tested on agar plates and results obtained in 24 hours, notes the lead author of the study, Nikos Fatsis-Kavalopoulos, who developed the method at Uppsala University.
The approach is based on creating a concentration gradient of antibiotics that have been cast into an agar plate, using a 3D-printed plastic disc. On the agar plate, bacteria that have been isolated from an individual patient are then cultured to see how they react to different combinations of antibiotics.
In their study, the researchers investigated E. coli bacteria isolated from urinary tract infections. Different cultures of E. coli proved not to react in the same way to specific antibiotic combinations. A combination of antibiotics that had synergistic effects on most cultures brought about antagonism in some, with the result that the treatment for the latter group was inferior.
“Antibiotic combination therapies are important for the efficient treatment of many types of infections, including those caused by antibiotic-resistant pathogens. Combination treatment strategies are typically used under the assumption that synergies are conserved across species and strains, even though recent results show that the combined treatment effect is determined by specific drug–strain interactions that can vary extensively and unpredictably, both between and within bacterial species,” write the investigators.
“To address this problem, we present a new method in which antibiotic synergy is rapidly quantified on a case-by-case basis, allowing for improved combination therapy. The novel CombiANT methodology consists of a 3D-printed agar plate insert that produces defined diffusion landscapes of 3 antibiotics, permitting synergy quantification between all 3 antibiotic pairs with a single test. Automated image analysis yields fractional inhibitory concentration indices (FICis) with high accuracy and precision.”
“A technical validation with 3 major pathogens, Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus aureus, showed equivalent performance to checkerboard methodology, with the advantage of strongly reduced assay complexity and costs for CombiANT. A synergy screening of 10 antibiotic combinations for 12 E. coli urinary tract infection (UTI) clinical isolates illustrates the need for refined combination treatment strategies.”
“For example, combinations of trimethoprim (TMP) + nitrofurantoin (NIT) and TMP + mecillinam (MEC) showed synergy, but only for certain individual isolates, whereas MEC + NIT combinations showed antagonistic interactions across all tested strains.”
“These data suggest that the CombiANT methodology could allow personalized clinical synergy testing and large-scale screening. We anticipate that CombiANT will greatly facilitate clinical and basic research of antibiotic synergy.”
“This result may be of great clinical importance. Consequently, instead of assuming that synergistic and antagonistic interactions are equal for all bacterial isolates, we test individually every isolate taken from an infected patient,” says Dan I. Andersson, PhD, professor of medical bacteriology at Uppsala University, who is primarily responsible for the study.
Customizing the drug combo in this way may be crucially important in achieving high efficacy in the treatment of infections, he continued, adding that being a simple, low-cost method, it is also easy to introduce and use in health care.


Comments are closed.