
Researchers have discovered soil in the West Fermanagh scarplands contains several species of antibiotic producing organisms.[Traditional Medicine Group]
One of the biggest threats to global health, food security and development, as recognized by the World Health Organization, is antibiotic resistance. The indiscriminate use of antibiotics since the discovery of penicillin in 1928, has led scientists to predict that antibiotic resistant microorganisms could kill up to 1.3 million people in Europe by 2050.
The urgent quest to discover new and chemically diverse antibiotics, especially against multi-resistant Gram-negative bacteria, that can be put to use once the current antibiotics lose efficacy is therefore of topmost priority, and has led scientists to explore unconventional sources, including folk medicine.
Findings in the article “The isolation of a novel Streptomyces sp. CJ13 from a traditional Irish folk medicine alkaline grassland soil that Inhibits multiresistant pathogens and yeasts,” published in the journal MDPI Applied Microbiology, report new sources of antibiotic discovery that refutes the widely held notion that original sources of antibiotics have been largely exhausted.
The study led by Gerry A. Quinn, PhD, scientist at the School of Biomedical Sciences, Ulster University, Northern Ireland, who grew up in Boho, shows soil used in ancient Irish folk medicine in the West Fermanagh scarplands contains several species of antibiotic producing organisms. This area of caves, alkaline grassland and bog is scattered with many remnants of Neolithic habitation.
An earlier analysis of the soil in this region led by the same team, had revealed an unknown strain of bacteria effective against four of the top six hospital infections that are resistant to antibiotics, including MRSA (Methicillin-resistant staphylococcus aureus).
The research team has shifted its focus since then to another region of the West Fermanagh scarplands that is also alkaline and associated to traditional folk medicine.
“Respecting the strong sensitivities around this specific soil, which remains within consecrated ground, we feel this cannot be considered as an exploitable resource for scientists in any normal sense. Instead, we returned to an adjacent upland alkaline grassland region which is still associated with traditional medicine approximately a kilometer from our original discovery to examine whether the same type of soil could be a new resource for antimicrobial discovery,” the authors note.
Paul Facey, PhD, one of the lead researchers from Swansea University says, “The fact that traditional medicine is incorporated in many local folk tales led us to believe that there was a good possibility of finding strong antibiotic producing organisms in other locations in these limestone hills.”
Soil samples from the new site revealed an even wider range of antimicrobial activity than the previous report. The DNA sequence of the new bacterial species has been deposited in the American national collection.
Antibiotic tests performed by Quinn and his colleagues reveal that the newly discovered bacterial species, Streptomyces sp. CJ13, inhibits the growth of several multi-resistant organisms, including Gram negative Pseudomonas aeruginosa—a common opportunistic pathogen associated with chronic lung infections in cystic fibrosis patients, MRSA—a common opportunistic pathogen often resistant to many antibiotics, anaerobic bacteria—linked to serious infections in deep wounds, and the yeast species, Candida.
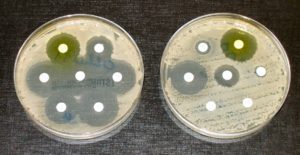
“We also examined the effects of heat and storage on the stability of the inhibitory substances present in the growth media of Streptomyces sp. CJ13,” the authors note.
Tests showed that the agar medium on which Streptomyces sp. CJ13 was grown retained its antibiotic activity even after 11 months of storage at 4–10 ⁰C, however the efficacy of the activity was reduced by about 40%.
Given the significant contributions made by Streptomyces to the fields of cancer and anti-viral therapies, team member Hamid Bakshi, PhD, says, “We are confident in the great potential of our most recent discovery to provide many interesting discoveries.”
Further studies will be required to establish the chemical identity and molecular structures of the new antibiotic compounds produced by the new Streptomyces species. However initial studies indicate that some genetic similarities exist between the genes of the new species and other antibiotic producing genes. The difference between the antibiotic genes found in Streptomyces sp. CJ13 and earlier antimicrobial genes suggests that these could be new variety of antibiotics.
The past few decades have witnessed a lull in antibiotic discovery. Discovery pipelines had depended largely on automated combinatorial chemical processes that screen large repositories of small compounds derived from antibiotic core structures against an array of bacterial pathogens. Major pharmaceutical companies have therefore largely abandoned investing in antibiotic discovery due to limited returns.

