While researchers have already succeeded in obtaining sequence information from single DNA strands, a further major challenge in bioanalytics is the direct determination of the amino acid sequence from individual proteins. Now a team from the University of Cergy-Pontoise in France, the University of Freiburg in Germany, and the University of Illinois in the United States, reports that they have been able to differentiate, for the first time, between individual amino acids in short peptides using a tiny pore the size of a nanometer.
The scientists, who published their study (“Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore”) in Nature Biotechnology, say they have thus laid the foundation for direct sequencing of individual proteins, i.e., via “nanopore protein sequencing.”
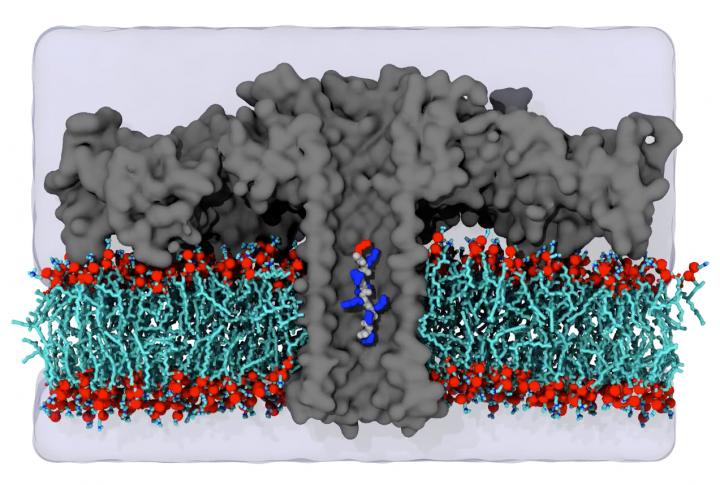
“This structural feature means that each amino acid spends sufficient time in the pore for sensitive measurement of the excluded volume of the amino acid. We show that distinct current blockades in wild-type aerolysin can be used to identify 13 of the 20 natural amino acids. Furthermore, we show that chemical modifications, instrumentation advances, and nanopore engineering offer a route toward identification of the remaining seven amino acids.”
Because previous techniques, such as mass spectrometry, are not sensitive enough to accurately determine the protein composition of a single cell, the pore-forming protein aerolysin was incorporated into an artificial cell membrane and electrodes were used to pass an ion current through the pore, explained Jan C. Behrends, MD, professor of physiology at the Institute of Physiology at the Medical Faculty of the University of Freiburg.
Several years ago, researchers at the Universities of Freiburg and Cergy demonstrated that blocking this current caused by a molecule entering the pore enabled the sensitive measurement of its size. On this basis, the team was able to show that the sensitivity of the so-called aerolysin pore is so high that short proteins, i.e. peptides, that only differ in a single amino acid can be distinguished from each other.
Using a high-resolution electrophysiological measurement method developed at the University of Freiburg, the team was able to differentiate between the amino acids leucine and isoleucine with more than 90% reliability. These two amino acids have the same composition, and therefore mass, and differ only in the spatial arrangement of the molecular groups.
The differentiation of such so-called structural isomers by means of the aerolysin pore proves that the current signal is not exclusively dependent on the molecular mass, as was previously assumed, thereby showing that the new technology is superior, in principle, to mass spectrometry, noted Behrends. The researchers in the United States used simulations to demonstrate that this high resolution is based on a kind of molecular trap within the pore. This trap immobilizes the peptides for about one hundredth of a second, which is what makes accurate measurement possible. This enabled the recently published study to reliably differentiate 11 of the 20 amino acids involved in the construction of proteins, using the nanopore current signal without additional chemical alterations.



