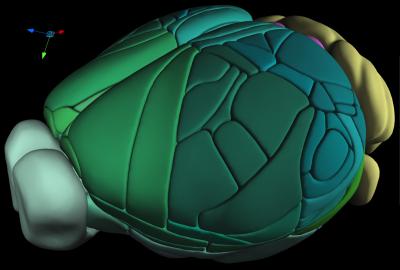
A team of scientists at the Allen Institute for Brain Science is making publicly available a manually constructed, 3D cellular-level atlas of the complete mouse brain. Derived from serial two-photon tomograph images of the brains of 1,657 mice, this third iteration of the Allen Mouse Brain Common Coordinate Framework, or CCFv3, is the result of three years of intensive data gathering and drawing.
“We hope the wider neuroscience community will use it as a new standard reference atlas,” said Lydia Ng, PhD, who is senior director of technology at the Allen Institute for Brain Science, and one of the senior authors on the atlas paper, which is published in Cell. “By making our atlas and related tools open access, new data and data types generated across our community can be more easily integrated and compared in the same spatial context, and the atlas, in turn, can be modified as our knowledge about brain structure evolves.”
Added co-senior author, Julie Harris, PhD, associate director of neuroanatomy at the Allen Institute for Brain Science, “Reference atlases are truly multi-purpose tools that are used for teaching neuroanatomy, providing common nomenclature to identify brain regions, supporting analyses to describe where data was collected, and representing our collective current knowledge on the organization of brain structure.” The researchers reported on the CCFv3 map in a report titled, “The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas.”
This video depicts a fusion of data in the CCF framework The background grayscale image represents the average anatomy of 1,675 individual specimens forming the basis for the common coordinate system. The colored curved lines represented sampled streamlines. The mouse cortex is a 3D sheet organized into layers where the connection between the layers is typically perpendicular to the surface, suggesting a hypothetical columnar organization. The curvature of the cortex makes it difficult to visualize along this theoretical dimension. These streamlines are an estimate of these “verticals” based on the curved geometry. To see if the streamlines reflect the true curvature we compared them with real data. The hotmetal colored image is a composite of multiple datasets to visualize the shape of thick-tufted dendrite of L5 pyramid neurons that were selectively labeled with Cre-dependent viral tracer injection into the Sim1-Cre_KJ18 or A930038C07Rik-Tg1-Cre driver line. Each dataset was registered to the CCF to allow the overlaying data from ~100 specimens. [Allen Institute for Brain Science]
The mouse brain contains approximately 100 million cells across hundreds of different regions. Knowledge of mouse brain neuroanatomy has advanced dramatically over the past decade, with the advent of whole-brain mapping projects adding new data types and revealing cellular architecture in rich new detail.
Large-scale international collaborations are generating major surveys of cell types and connections in the mouse brain, collecting large amounts of data across modalities, spatial scales, and brain areas, the authors explained. And as neuroscience datasets grow larger and more complex, a common spatial map of the brain becomes more critical, as does the ability to precisely co-register many different kinds of data into a common 3D space to compare and correlate.
“Recent large-scale international collaborations are generating major surveys of cell types and connections in the mouse brain, collecting large amounts of data across modalities, spatial scales, and brain areas,” the team commented. “Successful integration of these data requires a standard 3D reference atlas.” This means that modern digital reference atlases must evolve to stay current, Harris commented. “Yet classic standard atlases do not meet the demands of these cellular-resolution datasets in 3D. So, we produced a truly 3D atlas to serve as a standard anatomical framework for the whole-brain datasets being produced.”

“CCFv3 is parcellated into 43 isocortical areas and their layers, 329 subcortical gray matter structures, 81 fiber tracts, and 8 ventricular structures (per hemisphere),” the scientists explained. “Given that each dataset reveals a unique labeling pattern for certain mouse brain regions, combining all these data types should complement or confirm each other, demonstrating a tremendous methodological advantage for accurately defining brain structures,” Ng noted.
The authors say that CCFv3 has higher spatial resolution than any currently existing 3D mouse brain reference space based on magnetic resonance imaging. Because CCFv3 is an average from a large population of mice, it can be used to study inter-individual variability in the volumes of 3D brain structures in the context of different disease states. In addition, many anatomical details were apparent in the average brain that were not readily discernable in any single mouse brain.

“In the old days, people would define different regions of the brain by eye. As we get more and more data, that manual curation doesn’t scale anymore,” said Ng. “Just as we have a reference genome sequence, you need a reference anatomy.”
Historically, brain atlases were drawn in 2D, taking sheet-like views of the brain at different depths and lining them up. For some types of data, this form of brain mapping works well. But for modern neuroscience studies looking at neuron activity or cell characteristics across the entire brain, a 3D atlas gives better context. To make the atlas, the researchers broke up the brain into tiny virtual 3D blocks, known as voxels, and assigned each block a unique coordinate. The data that fed into that 3D construction came from the average brain anatomy of 1,675 different animals. The team then assigned each of those voxels to one of hundreds of different known regions of the mouse brain, drawing careful borders between distinct areas.

“We are now using this atlas as the common anatomical reference space for many large-scale projects, including whole-brain circuit mapping, single-cell reconstructions, and the generation of a comprehensive brain cell-type census,” Harris stated. “All these data, and what we learn about brain architecture in turn, will at some point necessitate an updated, next-generation 3D atlas, although it is likely to be constructed with different, more automated, data-driven methods.”
Future iterations of the atlas will thus likely rely on machine learning or other forms of automation, rather than on the laborious manual curation that went into the current version. “As we know now, atlases should be evolving and living resources, because as we learn more about how the brain is organized, we will need to make updates,” Harris said. “Building atlases in an automatic, unbiased way is where the field is likely moving.”
The Allen CCF is open access, and available with related tools at https://portal.brain-map.org/