Genome engineers have been using catalytically dead versions of CRISPR-Cas9 to interfere with the transcription of selected genes. Bacteria can do something similar—except that they can use CRISPR-Cas9 that is catalytically active. In other words, bacteria deploy CRISPR-Cas9 that is perfectly capable of snipping DNA but refrains from doing so. Instead, it looms over a gene, Scissors of Damocles style, inhibiting the gene’s expression.
In bacteria, CRISPR-Cas9 is best known for its role in immune defense, that is, cutting up foreign DNA that bacteriophages would use to infect their hosts. CRISPR-Cas9 also helps bacteria regulate endogenous genes, report scientists from Emory University School of Medicine and the Max Planck Unit for the Science of Pathogens. According to these scientists, the pathogenic bacterium Francisella novicida uses CRISPR-Cas9 to regulate the expression of genes essential to the bacterium’s virulence. No cutting is involved. Just obtrusive forbearance.
Details of this work appeared June 27 in the journal Molecular Cell, in an article titled, “Catalytically Active Cas9 Mediates Transcriptional Interference to Facilitate Bacterial Virulence.” The article describes the mechanism by which CRISPR-Cas9 controls a highly specific regulon of four virulence genes.
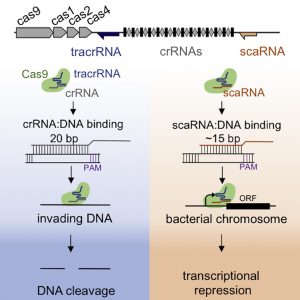
“Regulation occurs through a protospacer adjacent motif (PAM)-dependent interaction of Cas9 with its endogenous DNA targets, dependent on a non-canonical small RNA (scaRNA) and tracrRNA,” the article’s authors wrote. “The limited complementarity between scaRNA and the endogenous DNA targets precludes cleavage, highlighting the evolution of scaRNA to repress transcription without lethally targeting the chromosome.”
Emory microbiologist David Weiss, PhD, and colleagues had identified Cas9 several years ago when looking for genes that regulated F. novicida‘s virulence. F. novicida is a close relative of the bacterium that causes tularemia, and it grows inside mammalian cells. For the current paper elucidating why Cas9 is important for virulence, researchers in the lab led by Weiss teamed up with researchers in Germany led by Emmanuelle Charpentier, PhD, director at the Max Planck Institute for Infection Biology, whose work on the CRISPR/Cas9 nuclease led to its use as a gene editing tool.
The researchers determined that when CRISPR-Cas9 is acting to block gene activity, the nuclease uses a different kind of guide RNA sequence, a shorter guide RNA, one that allows targeted binding, but that doesn’t allow the nuclease to cut whatever DNA is targeted.
“We show that scaRNA can be reprogrammed to repress other genes,” the article’s authors added. In fact, the researchers were able to re-engineer CRISPR-Cas9 to repress a new target, a gene that makes the bacteria resistant to a last-line antibiotic, resensitizing the bacteria to antibiotic treatment. The researchers also demonstrated that “with engineered, extended complementarity to an exogenous target, the repurposed scaRNA:tracrRNA-FnoCas9 machinery can also direct DNA cleavage.”
According to the researchers, natural CRISPR-Cas9 transcriptional interference “likely represents a broad paradigm of regulatory functionality, one that may be critical to the physiology of numerous Cas9-encoding pathogenic and commensal organisms.” For example, CRISPR-Cas9 may help various kinds of bacteria cause disease.
“These findings raise the possibility that turning genes on and off may be a broad function of Cas9 in diverse bacteria,” noted graduate student Hannah Ratner, the first author of the Molecular Cell paper. “A question raised by this study is whether the ability of Cas9 to repress transcription can help explain the vast number of unidentified Cas9 targets.
“The programmability of the same protein for multiple different functions highlights and expands the incredible versatility of Cas9 for genome engineering applications.”


