June 15, 2013 (Vol. 33, No. 12)
An Effort on the Size and Scale of the HGP Is Needed to Figure Out What Variation Means
“Genome 2.0? Aren’t we on, like, Genome 1,000 by now?”
Yes, we are. In fact, more than 4,300 different whole genomes have been sequenced, nearly 200 of which are eukaryotic. Moreover, with targeted exome resequencing included, many more thousands of genomes have been analyzed for their coding sequence variations. Genetic information is no longer the bottleneck.
Don’t get me wrong, I’m not critiquing genome sequencers—quite the opposite, in fact. This amazing technology and related work is now defining the blueprint of life, its predicted functional units, and how it evolves and adapts.
In human populations, the full panoply of subtle variations that exist will soon be known, some of which will influence or control our morphological features, longevity, and predisposition to various diseases. Cancer, the prime example of our genetics going seriously awry, is now being defined in such great detail and complexity that many researchers (myself included) are increasingly optimistic that we will develop a sufficient range of targeted—or personalized—therapies and diagnostics to treat and detect cancers earlier and more effectively.
While such studies should continue, they are generating a new imperative to start to decipher what all this genetic information means. What does most of the genome—80% of which was recently defined by the ENCODE project to be functional in some way—do? What is normal genetic variation? Which variations will lead or contribute to pathological conditions? How do we validate which gene variations represent good targets for therapeutic intervention? Global genomics efforts now need to swing toward answering these key questions.
The magnitude of effort required to do so will likely dwarf the original efforts to sequence the first human genome. However, it’s not a mission we should shy away from, and just like the first Human Genome Project, it will take a concerted global collaborative effort to achieve it.
Post-genomic technologies are now maturing at a speed analogous to those seen with DNA sequencers, which will bring detailed functional genomic studies within every lab’s grasp.
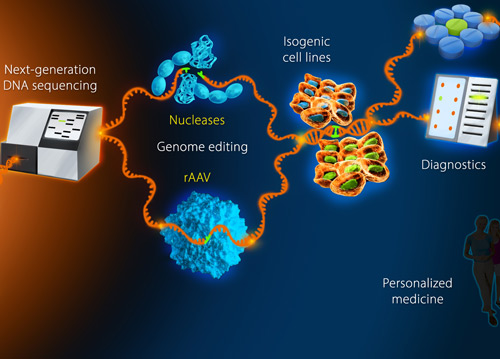
Bioinformatics and GWAS studies will continue to aid the deciphering process, triaging candidate functional units for further testing, but new facile methods to directly alter the endogenous genome of human cells growing in culture (previously only the domain of mouse geneticists and researchers studying lower organisms) will be essential to understanding what genes and genome variations functionally do.
It is still surprisingly underappreciated that robust and efficient targeted human genome editing techniques are now widely available to either knock out specific genes or loci of interest, or precisely recreate or correct disease causing variations in vitro to see how they alter cell biology.
One no longer needs to study genes using ectopic overexpression vectors, which in no way recapitulate normal gene function. Moreover, for the predominant nonprotein coding part of the human genome, with its long-range regulation and structure, extra-chromosomal plasmid-based systems will become virtually useless.
One should directly alter the genome itself in order to develop a deep and accurate understanding of its function. But what are the techniques available now and what can they do?
Genome editing techniques broadly fall into two groups: the programmable nucleases (zinc finger, TAL-effector, and most recently, CRISPR) and DNA-based homologous recombination vectors (of which the recombinant-AAV method is by far the most efficient in human and other mammalian cell-types). Each technology has its pros and cons.
Nucleases are adept at performing rapid multi-allelic knock-outs by cleaving DNA at a chosen locus and then allowing the error-prone nonhomologous end-joining (NHEJ) DNA-repair pathway to insert or delete bases at the break prior to re-joining the DNA-ends, thus leading to gene/loci disruptions. If more complex knock-ins or user-defined alterations are required, one can co-deliver a DNA-donor sequence harboring the desired change, which will recombine at the cleaved locus at a certain frequency. However, this knock-in process is significantly less efficient than for knock-outs.
Short, single-stranded DNA donors have recently been reported to increase the overall efficiency, but more complex or spatial alterations still remain a challenge due to the limitations of synthesizing longer DNA-oligos without errors. It should also be noted that there are concerns with nucleases generating off-target cleavage events; but, for performing relatively high-throughput targeted knockouts at low cost, this is probably not an overriding detraction.
CRISPR is a particularly exciting new entry into the nuclease field, as the Cas9 nuclease does not need redesigning to each locus of interest, but is programmable with a small RNA species with homology to the region in question. However, the number of genome sites accessible to CRISPR may be limited at this stage and the degree of off-target effects has not yet been determined. Its simplicity is very attractive and further advances are likely to be made.
In contrast to nucleases, the single-stranded DNA-virus rAAV uses the high-fidelity homologous recombination (HR) DNA-repair pathway exclusively to efficiently perform both knock-outs and knock-ins with precision (since no DNA cuts are required). However, because only one allele is usually modified at a time using rAAV, biallelic (or multiallelic) knock-outs will take longer than for nucleases. Nevertheless, the user-defined nature of the changes and the ability to perform longer-range and spatially separated alterations allows more complex inducible knock-out/knock-in strategies to be taken. Combined with the precision offered by rAAV, these are attractive features.
Further efficiency improvements will also likely be made for this technology and there are intriguing possibilities for combining nucleases with rAAV to enable the entire range of knock-ins and knock-outs to be brought into the high-throughput functional genomics arena.
With such a dawning capability, I believe the time is now right for governments, funding agencies and researchers to consider coming together to embark on determining the functional human genome. Let’s knock out every putative functional unit in the human genome to empirically understand what it does and modulate candidate SNPs suspected of disease involvement to unambiguously test whether they have cell-altering effects.
This Genome 2.0 effort would finally yield what we all naively hoped would be achieved by simply sequencing the human genome, but in fact was actually only the beginning. Who would like to join forces in finishing this journey?

Chris Torrance, Ph.D.
Chris Torrance, Ph.D. ([email protected]), is CSO at Horizon Discovery.