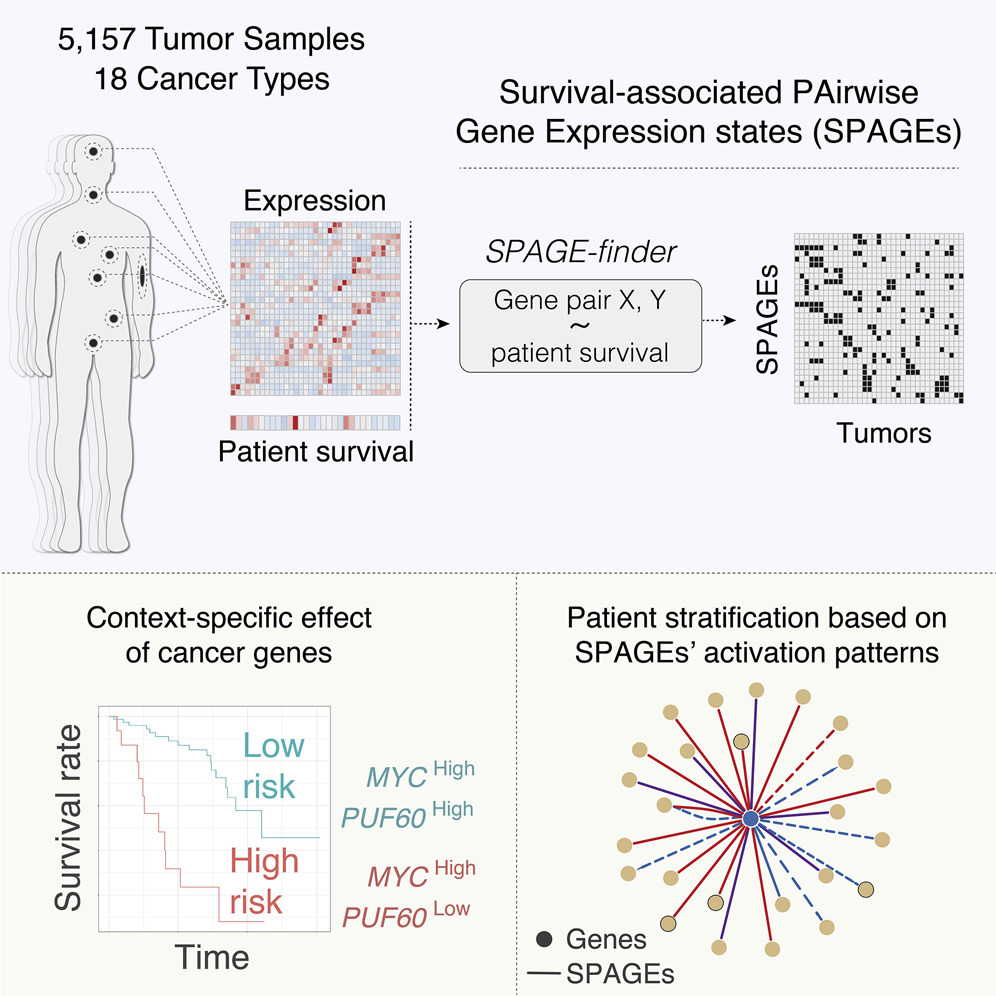
Genes do not normally work alone—on solo missions. Rather, they interact in highly cooperative networks with a perturbation of one gene affecting the expression of other genes. It is this teamwork that is ultimately responsible for the well being of a cell. Now, researchers have put tumor cell genes in the context of their interdependence. In doing so, they have identified 12 distinct types of gene-pair interactions in which varying levels of expression in the two genes correlate with cancer patient survival.
The paper, “Beyond Synthetic Lethality: Charting the Landscape of Pairwise Gene Expression States Associated with Survival in Cancer,” was recently published in Cell Reports.
Previous investigations of gene interactions in the context of cancer focused primarily on synthetic lethality (SL). Now, a team from the University of Maryland and the National Cancer Institute generalize this notion to identify 12 distinct gene-pair interactions in which varying levels of expression in the two genes correlated with cancer patient survival.
These important gene-pair relationships are in addition to synthetic lethality and many of these new relationships were more abundant in the researchers’ data than synthetic lethality, which means they may offer many more potential targets for cancer therapy.
“Our work expands the potential scope of strategies, thus far restricted to synthetic lethality, by generalizing the concept of exploiting genetic interactions to include many other yet unexplored types of gene-pair relationships,” said the study’s senior author Sridhar Hannenhalli, PhD, a professor in the department of cell biology and molecular genetics at the University of Maryland. “We believe this lays the foundation for using a computational method for identifying and studying additional types of genetic interactions in the future.”
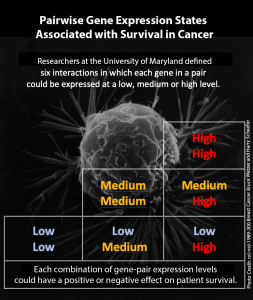
Researchers defined six interactions in which each gene in a pair could be expressed at a low, medium, or high level. They then considered that each of those combinations could be associated with a “positive” or “negative” outcome for patient survival. That brought the total number of potential gene-pair relationship types to 12. They analyzed data from 5,288 cancer tumors, representing 18 different cancer types.
Using a novel computational strategy, the researchers assessed all possible combinations of genes in their dataset—The Cancer Genome Atlas (TCGA.) Out of 163 million potential gene pairs, the researchers identified nearly 72,000 gene-pair interactions associated with a positive or negative patient survival. Of the genes involved in these interactions, a significant proportion are known to be involved in cell division and proliferation, which have clear links to cancer.
They define this more general concept of “survival-associated pairwise gene expression states” (SPAGEs) as gene pairs whose joint expression levels are associated with survival and describe their data-driven approach as SPAGE-finder. The authors write that the “detected SPAGEs explain cancer driver genes’ tissue specificity and differences in patients’ response to drugs and stratify breast cancer tumors into refined subtypes” and that these results “expand the scope of cancer SPAGEs and lay a conceptual basis for future studies of SPAGEs and their translational applications.”
Hannenhalli asserts that identifying gene-pair relationships can help scientists understand why mutations in certain genes lead to cancer in one tissue but not another, because their interacting partners might be expressed differently in different types of tissue. Similarly, gene-pair relationships could explain why certain drugs are effective for one patient but not another. The relationships also might help researchers identify subtypes of certain cancers, such as breast cancer, which may help with prognosis and therapy.
Using their findings on gene-pair interactions, the researchers were able to better predict patient outcomes in their data on tumor gene expression, compared with conventional methods that use expression of individual genes alone.
Hannenhalli stressed that there is still much work to be done to identify which gene pairs actually have a direct impact on cancer patient survival. The next step, he said, is to collaborate with cancer biologists or clinicians to begin experimenting with therapies targeted at some of the gene pairs identified in this study.