A new gene drive system introduces gears well suited to the transmission of relatively slight genetic variants, that is, allelic variants involving only one or a few base pairs. Typical gene drive systems are configured to copy relatively large DNA cargo sequences. The new gene drive system—or allelic drive system—favors or disfavors selected alleles and promises to accelerate the assembly of complex genotypes, which may consist of “point-mutant alleles in combination with insertional transgenes for a multitude or research and applied purposes,” said its creators.
The allelic drive comes from the laboratory of Ethan Bier, PhD, a professor of cell and developmental biology at the University of California San Diego (UCSD). According to Bier and colleagues, the allelic drive is equipped with a guide RNA (gRNA) that directs the CRISPR system to cut undesired variants of a gene and replace it with a preferred version of the gene. The new drive extends scientists’ ability to modify populations of organisms with precision editing.
In one example of its potential applications, specific genes in agricultural pests that have become resistant to insecticides could be replaced by original natural genetic variants conferring sensitivity to insecticides using allelic drives that selectively swap the identities of a single protein residue (amino acid).
In addition to agricultural applications, disease-carrying insects could be a target for allelic drives.
“If we incorporate such a normalizing gRNA on a gene-drive element, for example, one designed to immunize mosquitoes against malaria, the resulting allelic gene drive will spread through a population. When this dual action drive encounters an insecticide-resistant allele, it will cut and repair it using the wild-type susceptible allele,” said Bier. “The result being that nearly all emerging progeny will be sensitive to insecticides as well as refractory to malaria transmission.”
Details of the new system appeared April 9 in the journal Nature Communications, in an article titled, “Efficient allelic-drive in Drosophila.” The article describes how Bier’s team tested two configurations for allelic drive: copy-cutting, in which a nonpreferred allele is selectively targeted for Cas9/guide RNA (gRNA) cleavage, and a more general approach, copy-grafting, that permits selective inheritance of a desired allele located in close proximity to the gRNA cut site.
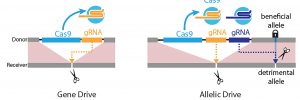
“Copy-cutting involves a Cas9-gRNA complex selectively cutting one allelic variant, followed by homology-directed repair (HDR)-mediated repair and replacement with a noncleavable allele of the same gene provided in trans,” the article’s authors wrote. “The second, and more generally applicable form of allelic drive, which we refer to as copy-grafting, involves copying a short genomic interval that encompasses a favored allele in proximity to a gRNA cut site. In the case of copy-grafting, the favored allele is associated with neighboring sequences resistant to gRNA cleavage.”
Although demonstrated in fruit flies, the new technology also has potential for broad application in insects, mammals, and plants. According to the researchers, several variations of the allelic drive technology could be developed with combinations of favorable traits in crops that, for example, thrive in poor soil and arid environments to help feed the ever-growing world population. Beyond environmental applications, allelic drives should enable next-generation engineering of animal models to study human disease as well as answer important questions in basic science.
“Forcing species to return to their natural sensitive state using allelic drives would help break a downward cycle of ever-increasing and environmentally damaging pesticide over-use,” noted Annabel Guichard, PhD, a project scientist in the Bier lab at UCSD and the paper’s first author.
“An unexpected finding from this study is that mistakes created by such allelic drives do not get transmitted to the next generation,” she added. “These mutations instead produce an unusual form of lethality referred to as ‘lethal mosaicism.’ This process helps make allelic drives more efficient by immediately eliminating unwanted mutations created by CRISPR-based drives.”
Lethal mosaicism dominantly eliminates NHEJ-induced mutations and favors inheritance of functional cleavage-resistant alleles.
The Bier team indicated that their allelic drive methods, enhanced by lethal mosaicism and a trans-generational drive process they refer to as shadow drive, could have “broad practical applications in improving health and agriculture and greatly extend the active genetics toolbox.”


