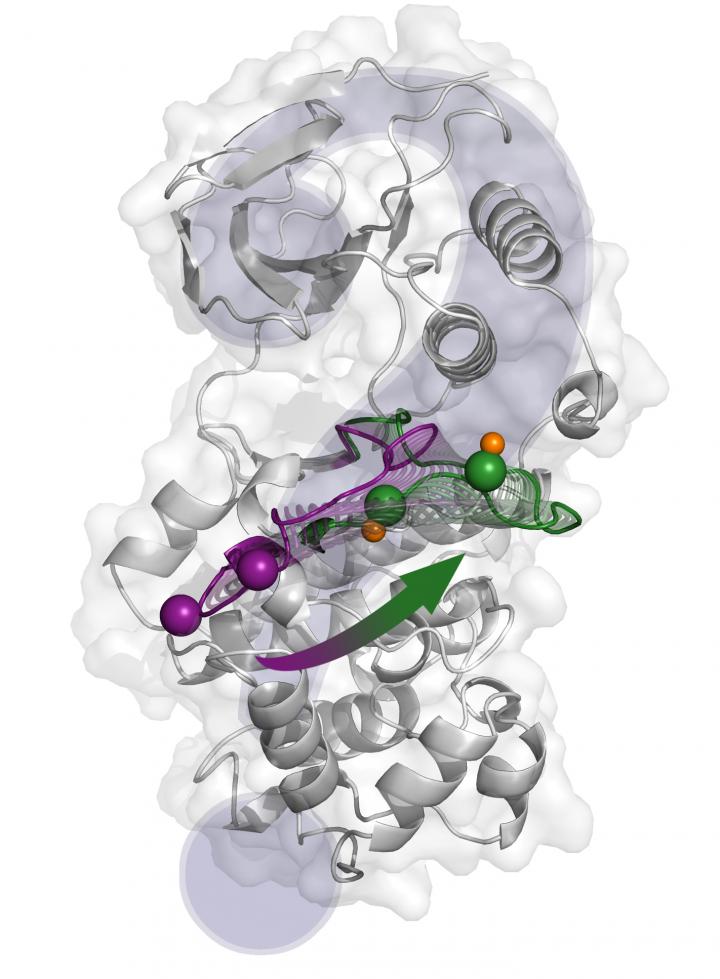
![The structural changes from the inactive state (purple) to the active one (green) proposed by X-ray crystallography. [Antonija Kuzmanic/IRB Barcelona]](https://genengnews.com/wp-content/uploads/2018/08/Apr28_2017_AntonijaKuzmanic_p38ActivationMechanism2122211212-1.jpg)
The structural changes from the inactive state (purple) to the active one (green) proposed by X-ray crystallography. [Antonija Kuzmanic/IRB Barcelona]
Scientists at the Institute for Research in Biomedicine in Barcelona have published a study that provides an integrative picture of the p38α activation mechanism and offers new insights into the molecular effects of various molecules that regulate the enzymatic activity of this protein.
The p38α protein is a member of a family of molecules that transmit outside signals throughout the cell, thus allowing for an appropriate cell response, such as proliferation, differentiation, senescence, or death. Moreover, the participation of p38α in pathological conditions, like chronic inflammatory diseases and cancer, makes it a promising pharmacological target. In this regard, a complete picture of the activation mechanism of this protein is essential in order to design specific inhibitors that do not affect other processes.
Using computational techniques, researchers deciphered the key elements of the complex molecular mechanism underlying p38α activity. They say their paper (“Changes in the Free-Energy Landscape of p38α MAP Kinase through Its Canonical Activation and Binding Events as Studied by Enhanced Molecular Dynamics Simulations”), which appears in eLife, describes the protein activation mechanism in unprecedented detail and reconciles the apparent contradictory results reported in previous structural studies.
“Considering the importance of p38α for pathological processes, we hope the knowledge obtained in this study will help to target the protein with more specificity,” noted Antonija Kuzmanic, Ph.D., first author of the study and an EU Marie Curie COFUND fellow.
p38α has already been targeted for inflammatory diseases and some types of cancer. However, none of the drugs has yet made it to the market. “Our study reveals novel conformations of the protein, which could be used as a starting point in virtual screening studies aimed at uncovering new inhibitors,” explained Dr. Kuzmanic, adding that the team was also able to highlight important electrostatic interactions, which may allow the investigators to explore alternative activation pathways with increased specificity.
“We used only computational techniques. Mainly, we employed numerous molecular dynamics simulations combined with an advanced sampling technique called metadynamics,” she continued, noting that this combination has an advantage over standard molecular dynamics simulations, as it allows researchers to observe large conformational changes in a reasonable amount of computational time.
“We are able to add statistical significance to the conformations we observed in our simulations,” she said.