Anti-CRISPR proteins could be harnessed to improve CRISPR gene editing systems by reducing their off-target effects, and by controlling when and where they act. Anti-CRISPR proteins, however, are rare in nature—or simply hard to find. In hopes of adding to the few anti-CRISPR proteins already discovered, scientists based at the Technical University of Denmark tried a new kind of search. In doing so, they may have avoided the streetlight effect, a kind of observational bias that occurs when people search for something only where it seems obvious to look.
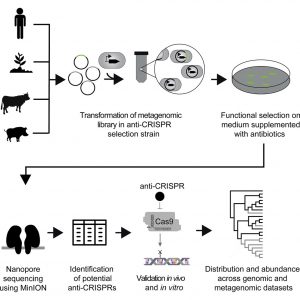
Instead of a streetlight’s illumination limiting a search for lost keys, for example, the usual anti-CRISPR search strategy—examining the contents of phage DNA—could hamper the search for CRISPR inhibitors. So, rather than use the usual strategy, the scientific team at the Technical University of Denmark used a genetic circuit to select anti-CRISPR genes from metagenomes. The team, led by Morten O.A. Sommer, who is also affiliated with the Novo Nordisk Foundation Center for Biosustainability, succeeded in discovering four CRISPR-Cas9 inhibitors.
Detailed findings appeared February 5 in Cell Host & Microbe, in an article titled, “Discovery and Characterization of Cas9 Inhibitors Disseminated across Seven Bacterial Phyla.” The article described how a synthetic genetic circuit enabled a high-throughput investigation of anti-CRISPR genes from metagenomic libraries. This investigation, the article emphasized, was based on functional activity rather than sequence homology or genetic context.
“We identified 11 DNA fragments from soil, animal, and human metagenomes that circumvent Streptococcus pyogenes Cas9 activity in our selection strain,” the article’s authors wrote. “Further in vivo and in vitro characterization of a subset of these hits validated the activity of four anti-CRISPRs.”
The DNA was chopped into smaller pieces and randomly expressed on a plasmid within a bacterial cell. This cell contained a genetic circuit for selection of anti-CRISPR activity. In short, this meant that cells containing a plasmid with a potential anti-CRISPR gene would become resistant to a certain antibiotic. On the contrary, cells in which the plasmid did not confer anti-CRISPR-activity would die. With this system, the researchers could easily detect and select DNA with anti-CRISPR activity and trace it back to its origin.
Further characterization could then confirm the activity of four new anti-CRISPRs. Phylogenetic analysis revealed that the genes identified in the fecal samples are present in bacteria found in multiple environments, for instance in bacteria living in insects’ gut, seawater, and food. This shows that the newly discovered genes are spread in many bacterial branches in the tree of life, and in some cases with evidence that some of these genes have been horizontally transferred numerous times during evolution.
These findings suggest that anti-CRISPRs could likely play a much bigger role in the interplay between phage and host than what has previously been suggested.
Earlier studies in this field have demonstrated that anti-CRISPR proteins can be used to reduce errors, such as cutting DNA at off-target sites, when doing genome editing in the laboratory.
“Today, most researchers using CRISPR-Cas9 have difficulties controlling the system and off-target activity. Therefore, anti-CRISPR systems are very important, because you want to be able to turn your system on and off to test the activity. Therefore, these new proteins could become very useful,” Sommer noted.
The researchers actually discovered that the four new anti-CRISPR proteins seem to have different traits and properties. Going forward, this will be very exciting to investigate further.