
Researchers in Korea have harnessed two complementary sequencing techniques to better understand the genetic architecture of the SARS-CoV-2 genome. By combining nanopore-based direct RNA sequencing (DRS) sequencing, and DNA nanoball (DNB) sequencing, the scientists, led by Narry Kim, PhD, and Hyeshik Chang, PhD, at the Center for RNA Research within the Institute for Basic Science (IBS) in Seoul, generated new insights into subgenomic RNAs (sgRNAs) that are translated into viral proteins. Analyzing the sequence information of each RNA also revealed where the genes are located on the long viral genomic RNA.

“Not only detailing the structure of SARS-CoV-2, we also discovered numerous new RNAs and multiple unknown chemical modification on the viral RNAs,” Kim commented. Our work provides a high-resolution map of SARS-CoV-2. This map will help us understand how the virus replicates and how it escapes the human defense system.” Findings by the Center for RNA Research team and collaborators at the Korea National Institute of Health (KNIH), within the Korea Centers for Disease Control & Prevention (KCDC), are available in a preprint of their paper, which is to be published in Cell, titled, “The architecture of SARS-CoV-2 transcriptome.”
SARS-CoV-2 is an enveloped virus with a positive-sense, single-stranded RNA genome of ~30 kb, the authors explained. SARS-CoV-2 belongs to the genus betacoronavirus, together with SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), with which it shares 80% and 50% homology, respectively. Coronaviruses carry the largest genomes—26–32 kb—among all the RNA virus families. When it infects host cells, SARS-CoV-2 replicates its genomic RNA (gRNA) and produces many smaller RNAs known as subgenomic RNAs. These sgRNAs are used for synthesizing various proteins. “The gRNA is packaged by the structural proteins to assemble progeny virions,” the researchers commented. ”Shorter sgRNAs encode conserved structural proteins, (spike protein (S), envelope protein (E), membrane protein (M), and nucleocapsid protein (N)), and several accessory proteins.”
Although recent studies have reported the sequence of the RNA genome, such studies were only able to predict where the genes might be. “For the development of diagnostic and therapeutic tools and the understanding of this new virus, it is critical to define the organization of the SARS-CoV-2 genome,” the team continued. For their newly reported work the researchers combined the two complementary sequencing techniques, nanopore direct RNA sequencing, and then DNA nanoball sequencing based on the sequencing by synthesis by synthesis principle (DNBseq), to further understand the genomic RNA and sgRNA architecture.
Nanopore-based direct RNA sequencing enables direct analysis of the entire, long viral RNA without fragmentation, the team noted. “While nanopore DRS is limited in sequencing accuracy, it enables long read sequencing, which would be particularly useful for the analysis of long nested CoV transcripts.” Conventional RNA sequencing methods usually require a step-by-step process of cutting and converting RNA to DNA before reading RNA, they pointed out. “And because DRS detects RNA instead of cDNA, RNA modification data can be obtained directly during sequencing.” In contrast, while the DNA nanoball sequencing technology can read only short fragments, it has the advantage of analyzing a large number of sequences with high accuracy. The two techniques used together provided detailed new information on the SARS-CoV-RNA genome.
While it was previously known that there are 10 subgenomic RNAs making up the viral particle structure, the research team’s studies identified open reading frames for nine subgenomic RNAs, but invalidated one of the open reading frames (ORFs). “It is important to note that ORF10 is represented by only one read in DNB data … and that it was not supported at all by DRS data,” they wrote. This ORF also showed no significant homolog to known proteins. “Thus, ORF10 is unlikely to be expressed,” they continued. “Taken together, SARS-CoV-2 expresses nine canonical sgRNAs (S, E, M, N, 3a, 6, 7a, 7b, 8) together with the gRNA.”
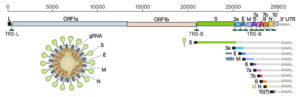
The investigators also found that there are dozens of unknown subgenomic RNAs, resulting from RNA fusion and deletion events. “DNA nanoball sequencing shows that the transcriptome is highly complex owing to numerous discontinuous transcription events,” they stated. “In addition to the canonical genomic and nine subgenomic RNAs, SARS-CoV-2 produces transcripts encoding unknown ORFs with fusion, deletion, and/or frameshift. Using nanopore direct RNA sequencing, we further find at least 41 RNA modification sites on viral transcripts, with the most frequent motif.”
Kim noted, “Though it requires further investigation, these molecular events may lead to the relatively rapid evolution of coronavirus. Moreover, we find multiple unknown chemical modifications on the viral RNAs. It is unclear yet what these modifications do, but a possibility is that they may assist the virus to avoid the attack from the host.”
The research team suggests that modified RNAs may have new properties that are different from unmodified RNAs even though they have the same genetic information in terms of RNA base sequence. They believe that if they can work out the unknown characteristics of RNA, they may discover new potential avenues for combatting SARS-CoV-2. “Unambiguous mapping of the expressed sgRNAs and ORFs is a prerequisite for the functional investigation of viral proteins, replication mechanism, and host-viral interactions involved in pathogenicity,” the scientists stated.
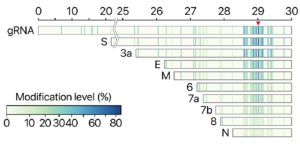
Newly discovered chemical modifications could also help to understand the life cycle of the virus. “Like other RNA viruses, CoVs undergo frequent recombination, which may allow rapid evolution to change their host/tissue specificity and drug sensitivity,” they pointed out. “Functional investigation of the unknown transcripts and RNA modifications discovered in this study will open new directions to our understanding of the life cycle and pathogenicity of SARS-CoV-2 … Our data provide a rich resource and open new directions to investigate the mechanisms underlying the pathogenicity of SARS-CoV-2.”
“Now we have secured a high resolution gene map of the new coronavirus that guides us where to find each bit of genes on all of the total SARS-CoV-2 RNAs (transcriptome) and all modifications RNAs (epitranscriptome),” Kim stated. “It is time to explore the functions of the newly discovered genes and the mechanism underlying viral gene fusion. We also have to work on the RNA modifications to see if they play a role in virus replication and immune response. We firmly believe that our study will contribute to the development of diagnostics and therapeutics to combat the virus more effectively.”



Comments are closed.