An investigator sifts through trash, uncovers clues, and assembles a case-cracking profile. Are we watching a crime drama? No, we’re following the progress of a scientific study, one that reaches into the cell’s trash and finds tell-tale patterns of protein turnover and degradation. In this case, the cells are immune cells, from patients suffering systemic lupus erythematosus (SLE), and the patterns are protein degradation signatures. These signatures, it happens, show what conventional protein expression signatures don’t. In fact, the trash-derived signatures point to a new understanding of SLE.
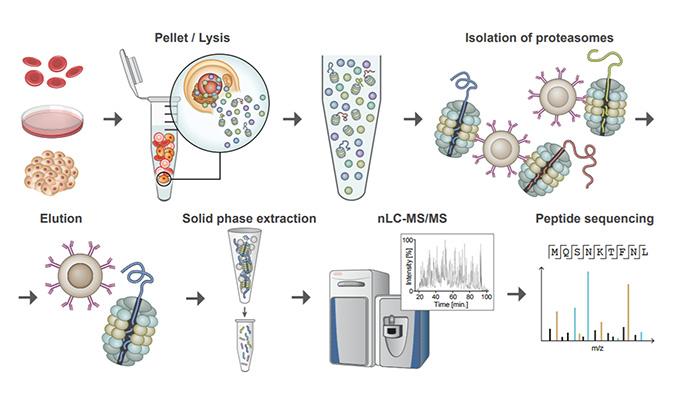
Just as important, the study demonstrates a cellular dumpster diving technique that might be useful in other cases of autoimmunity, as well as in cases of protein aggregation diseases and cancer. The technique, called Mass spectrometry Analysis of Proteolytic Peptides (MAPP), was developed by scientists based at the Weizmann Institute of Science, in the laboratory of principal investigator Yifat Merbl, Ph.D. It is a kind of proteomic fingerprinting method that allows for capture, isolation and analysis of proteasome-cleaved peptides.
“Current proteomics,” Dr. Merbl explains, “generates detailed lists of the proteins expressed in cells, which is like trying to understand the lifestyle of people by looking at successive snapshots of their house. We can get some sense of their taste in furniture and art, their ages, and maybe whether they like to read or cook.
“Getting successive snapshots of their trash, on the other hand, will tell us what they eat, what medications they take, what they buy and where they travelled. In the same way, when we looked at the contents of cellular trash cans, we were able to profile the disease state on the level of the cell’s activities and its protein turnover, as well as uncovering some clues as to what goes wrong in these cells to bring about the symptoms of the disease.”
Dr. Merbl’s team generated MAPP profiles of discarded proteins from healthy subjects and MAPP profiles of discarded proteins from lupus patients. When the investigators compared the profiles, they found an abnormally high turnover of particular proteins in the histone family. Histones have several vital functions within the cell, but they have been associated with disease, particularly autoimmune disease, when they are found in above-average quantities outside the cells.
Details of this forensic work appeared October 22 in the journal Nature Communications, in an article entitled, “Revealing the cellular degradome by mass spectrometry analysis of proteasome-cleaved peptides.” This article describes how the MAPP technique could be used to scrutinize the some 70% of the proteins in the body that are broken down by proteasomes, which are the cell’s degradation and recycling units.
“Application of MAPP … revealed dynamic modulation of the cellular degradome in response to various stimuli, such as proinflammatory signals,” the article’s authors wrote. “Further, we performed analysis of minute amounts of clinical samples by studying cells from the peripheral blood of patients with SLE.”
The current understanding says that in lupus, an excess of white blood cells die, spilling their contents, including the histones, into the bloodstream. The immune system then identifies the added histone peptides as foreign antigens and undertakes its response against these particular pieces of protein. “But our findings suggest that something takes place earlier in the white blood cells,” said Dr. Merbl. “These are cells that normally take peptides from bacteria or viruses and present them on their outer membrane like little flags alerting the immune system to a pathogen. We think the cells from the lupus patients might do the same thing with the extra histone peptides, which may be degraded due to an aberration in the proteins, and may then be identified as antigens by the rest of the immune system.”
Dr. Merbl also pointed out that the experiments were conducted on liquid biopsies from standard blood samples extracted from patients in the clinic, so if it can be shown in further experiments that the signature the group discovered is unique to lupus among similar autoimmune diseases, such testing could conceivably make its way to hospital labs in the not-so-distant future. The research group believes the level of detail they obtained through the new approach and technology may provide the resolution needed to help understand the underlying mechanisms of numerous diseases involving damaged proteins and the cellular proteasome system.
What is exciting to Merbl and her group, however, is the possibilities the MAPP method opens up: Since every cell has its own contingent of proteasomes for breaking down used-up and damaged proteins, the cellular “trash collection” technique could yield valuable insight into numerous aspects of basic and translational biology ranging from cancer and cancer immunotherapy, to basic research on protein regulation.
Because the proteins of the degradome are less abundant than structural proteins, standard proteomics surveys tend to miss them. But the degradome consists of “precisely the proteins that are crucial to the cell’s functions,” insisted Dr. Merbl, “and thus their malfunctioning plays a role in many diseases.


