The SKI complex, a protein complex involved in various RNA metabolism pathways, has been identified as a potential target for broad-spectrum antiviral drugs. Already, chemicals have been identified that interact with the SKI complex. Encouragingly, these chemicals have been shown to prevent the replication of coronaviruses, influenza viruses, and filoviruses in mammalian cell culture.
The SKI complex was originally identified in yeast, but it is also found in mammals. Both these SKI contexts figured in a recent search for broad-spectrum, host-directed, antiviral targets. The search, conducted by scientists at the University of Maryland School of Medicine (UMSOM), began with yeast suppressor screening, the intent of which was to find a functional genetic interaction between viral proteins and eukaryotic proteins.
Screens were conducted with influenza A virus (IAV) and Middle East respiratory syndrome coronavirus (MERS-CoV). The eukaryotic proteins of interest were those potentially involved in supporting viral replication and survival.
The screens suggested that the SKI was needed by both viruses.
Next, conducting experiments in mammalian systems, the UMSOM scientists determined that siRNA-mediated knockdown of SKI genes inhibited replication of IAV and MERS-CoV. This finding prompted the UMSOM scientists to consider how the SKI complex could be targeted by potential antiviral agents.
Details of this work appeared in the Proceedings of the National Academy of Sciences (PNAS), in an article titled, “The SKI complex is a broad-spectrum, host-directed antiviral drug target for coronaviruses, influenza, and filoviruses.”
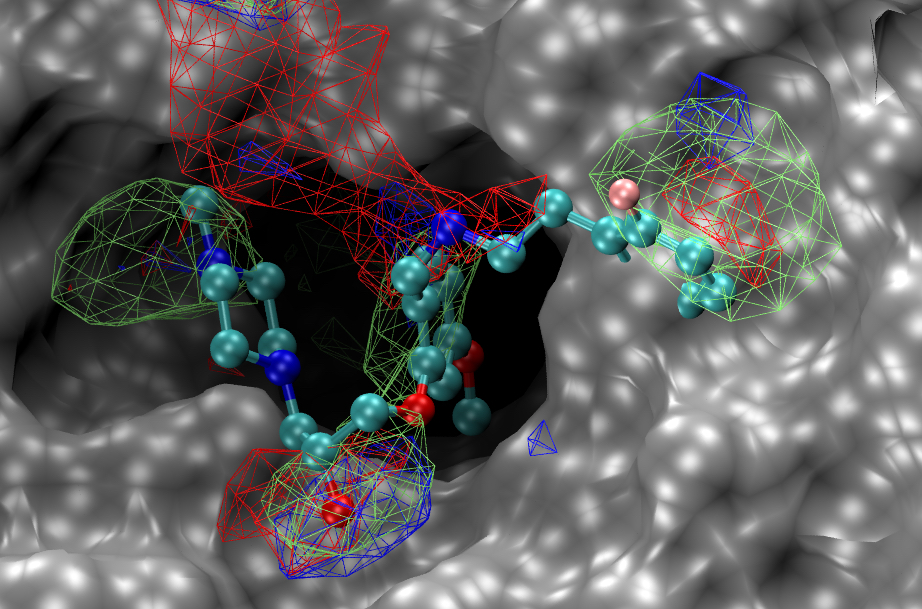
“In silico modeling and database screening identified a binding pocket on the SKI complex and compounds predicted to bind,” the article’s authors wrote. “Experimental assays of those compounds identified three chemical structures that were antiviral against IAV and MERS-CoV along with the filoviruses Ebola and Marburg and two further coronaviruses, SARS-CoV and SARS-CoV-2.”
The chemicals’ antiviral activity, the authors suggested, occurs through inhibition of viral RNA production.
“We determined that disrupting the SKI complex keeps the virus from copying itself, which essentially destroys it,” said Matthew Frieman, PhD, the study’s corresponding author and associate professor of microbiology and immunology at the UMSOM. “We also identified compounds that targeted the SKI complex, not only inhibiting coronaviruses but also influenza viruses and filoviruses, such as the one that causes Ebola.”
Frieman and his colleagues from the School of Pharmacy’s Computer-Aided Drug Design Center and the Center for Biomolecular Therapeutics at the UMSOM used computer modeling to identify a binding site on the SKI complex and identified chemical compounds that could bind to this site. Subsequent experimental analysis showed these compounds to have antiviral activity against coronaviruses, influenza viruses, and filoviruses (such as Ebola). Researchers from the National Institute of Allergy and Infectious Diseases also participated in this study.
The study was funded by Emergent BioSolutions, a biopharmaceutical company based in Gaithersburg, MD.
“These findings present an important first step in identifying potential new antivirals that could be used to treat a broad number of deadly infectious diseases,” said Stuart Weston, PhD, the study’s lead author and a research fellow at the UMSOM. Such drugs have the potential to treat infectious diseases associated with future pandemics. The next steps include conducting animal studies to learn more about the safety and efficacy of these experimental compounds, which are not approved by the FDA.
“At this stage, our compounds that display broad antiviral activity are only modeled to interreact with the SKI complex,” the article’s authors noted. “We find that loss of the SKI complex through siRNA targeting removes the antiviral activity of [one of our compounds], suggesting a requirement for the complex to produce the observed antiviral activity. Future work is aimed at further analyzing whether the compounds bind directly to the SKI complex using structural biology approaches and investigating whether other roles of the SKI complex are impacted.”