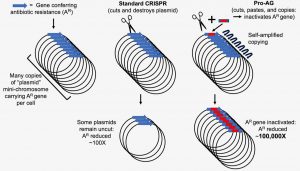
Where a gene drive that cuts and destroys plasmids may fail, a gene drive that cuts, pastes, and copies plasmids may succeed. The “where,” in this case, is an antibiotic-resistant bacterium that carries multiple copies of an antibiotic-resistance gene. That is, the target gene occurs on multiple copies of an “amplified” plasmid.
If some plasmids remain intact, the antibiotic-resistance gene persists and may even be passed to other bacteria. To overcome this problem, scientists based at the University of California (UC), San Diego, developed a gene drive that incorporates a self-amplifying mechanism. Called Pro-AG, for “proactive” active genetics, it increases its efficiency through a positive feedback loop.
Pro-AG was introduced December 16 in Nature Communications, in an article titled, “A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus.”
“Here, we develop an analogous CRISPR-based gene-drive system for the bacterium Escherichia coli that efficiently copies a gRNA cassette and adjacent cargo that are flanked with sequences homologous to the targeted gRNA/Cas9 cleavage site. This “proactive” genetic system functionally inactivates an antibiotic resistance marker on a high copy number plasmid with ~100-fold greater efficiency than control CRISPR-based methods, suggesting an amplifying positive feedback loop due to increasing gRNA dosage.”
The article’s authors, scientists in the laboratories of Victor Nizet, MD, and Ethan Bier, PhD, suggest that their Pro-AG system may reduce one of society’s most formidable threats to human health. Widespread prescriptions of antibiotics and use in animal food production have led to a rising prevalence of antimicrobial resistance in the environment.
Evidence indicates that these environmental sources of antibiotic resistance are transmitted to humans and contribute to the current health crisis associated with the dramatic rise in drug-resistant microbes. Health experts predict that threats from antibiotic resistance could drastically increase in the coming decades, leading to some 10 million drug-resistant disease deaths per year by 2050 if left unchecked.
The core of Pro-AG features a modification of the standard CRISPR-Cas9 gene editing technology in DNA. Pro-AG works by a cut-and-insert repair mechanism to disrupt the activity of the antibiotic-resistant gene with at least two orders of magnitude greater efficiency than current cut-and-destroy methods.
The study’s lead authors, Andrés Valderrama, PhD, postdoctoral fellow and Surashree Kulkarni, PhD, postdoctoral scholar, demonstrated the effectiveness of the new technique in experimental cultures containing a high number of plasmids carrying genes known to confer resistance to the antibiotic ampicillin. The system relies on a self-amplifying “editing” mechanism that increases its efficiency through a positive feedback loop. The result of Pro-AG editing is the insertion of tailored genetic payloads into target sites with high precision.
While Pro-AG is not yet ready for treating patients, “a human delivery system carrying Pro-AG could be deployed to address conditions such as cystic fibrosis, chronic urinary infections, tuberculosis, and infections associated with resistant biofilms that pose difficult challenges in hospital settings,” said Nizet, distinguished professor of pediatrics and pharmacy and the faculty lead of the UC San Diego Collaborative to Halt Antibiotic-Resistant Microbes (CHARM).
When combined with a variety of existing delivery mechanisms for spreading the Pro-AG system through populations of bacteria, the scientists say the technology also could be widely effective in removing, or “scrubbing,” antibiotic-resistant strains from the environment in areas such as sewers, fish ponds, and feedlots. Because Pro-AG “edits” its targets rather than destroys them, this system also enables engineering or manipulating bacteria for a broad range of future biotechnological and biomedical applications rendering them harmless or even recruiting them to perform beneficial functions.
“The highly efficient and precise nature of Pro-AG should permit a variety of practical applications, including dissemination of this system throughout populations of bacteria using one of several existing delivery systems to greatly reduce the prevalence of antibiotic resistance in the environment,” added Bier, a distinguished professor in the section of cell and developmental biology and science director of the UC San Diego unit of the Tata Institute for Genetics and Society (TIGS).
“The high efficiencies of accurate Pro-AG cassette copying in bacteria are comparable to those attained by gene-drive systems in diploid organisms (>90%),” the article’s authors concluded. “In addition to its high efficiency, an important feature of Pro-AG is the ability to perform precise and potentially subtle edits of a target gene rather than simply eliminating the target sequence or bacteria carrying that locus.”


