September 1, 2010 (Vol. 30, No. 15)
Technique Reveals Wide Range of Critical Changes in Real Time and Great Detail
Live-cell imaging, which has proven to be tremendously beneficial in helping scientists understand how cells work, is not without its wrinkles. “Live-cell imaging is key to understanding cellular differentiation and function,” said Magnus Persmark, Ph.D., senior product manager at Life Technologies, “yet cell types like stem cells, primary cells, and neurons have traditionally been quite challenging to label efficiently and without cytopathic effects.”
Another challenge, noted Anna Christensen, Ph.D., imaging product manager at Caliper Life Sciences, is specificity. “In vivo challenges to cell imaging, particularly targeting specific processes within the whole cell, remain,” she said. “When you look at a specific pathway in a specific animal, there are a lot of nonspecific events occurring. The real challenge lies in targeting relevant events and processes, in the cell, in the pathways, and in the whole animal.”
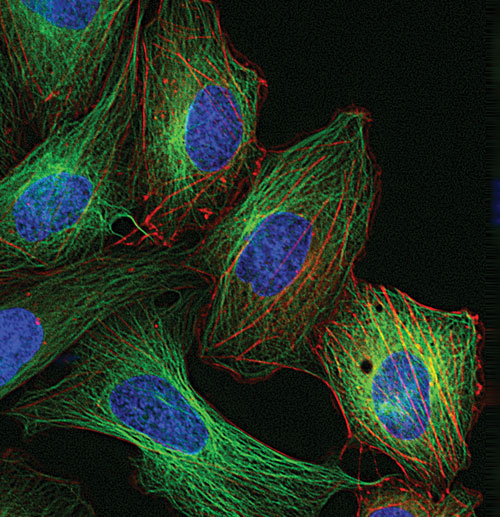
Live HeLa cells transduced with CellLight™ MAP4-GFP and CellLight Actin-RFP then labeled with Hoechst 33342 [Life Technologies]
At the “Focus on Microscopy” and “Immunology” meetings held earlier this year, investigators and vendors explored some of the issues facing researchers working with live-cell imaging and offered some solutions to help enhance research efforts.
The evolution of superresolution microscopy has been fairly rapid over the last few years and has lead to several important findings, particularly by using stimulated emission depletion (STED) microscopy, said Tanjef Szellas, product manager, superresolution marketing, Leica Microsystems.
“The implementation of continuous wave (CW) lasers emitting in the visible spectral range has opened up new perspectives for biomedical research,” Dr. Szellas noted.
STED microscopy uses the nonlinear de-excitation of fluorescent dyes to surmount the resolution limit imposed by diffraction with standard confocal laser-scanning microscopes.
The resolution of a confocal laser-scanning microscope is limited to the spot size to which the excitation spot can be focused. However, within the STED microscope, the diffraction limit is surmounted by targeted strong de-excitation of dye molecules, switching them off effectively.
Dr. Szellas discussed the capabilities of Leica’s commercial STED microscope, the TCS STED CW. “The integration of an orange CW laser to increase the resolution has made it possible to investigate fixed and living intact specimens labeled with standard fluorophores like Alexa488 and Oregon Green and also with fluorescent proteins like YFP.”
Dr. Szellas noted that there are quite a few applications where continuous wave stimulated emission depletion microscopy has been used, including cell structure research, vesicle trafficking, and neurobiology, particularly in the areas of synapse assembly and vesicle fusion, and to better understand structures of nuclear pore complexes.
“In general, it can be used anywhere where the following is needed: sub-80 nm optical resolution in an intact specimen—inside a cell, for example—with recording speed of up to 20 frames per second, especially for viruses and vesicles that exhibit average sizes of approximately 50–100 nm. To follow their exo- and endocytosis in real time with “real” sizes requires a set-up like STED.”
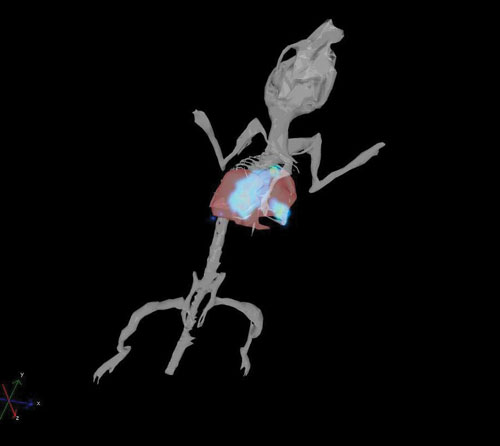
Reconstructed 3-D bioluminescent CT26-luc tumor in the lungs [Caliper Life Sciences]
Structured Illumination Microscopy
Another commonly used tool for live-cell imaging is wide-field optical microscopy, but there are some limitations for this method as well, noted Lin Shao, research specialist at the Howard Hughes Medical Institute’s Janelia Farm Research Campus. “The main problem is that its resolution is limited by the visible light’s wavelength and the objective’s aperture.”
Also referred to as wide-field structured illumination (SI), this approach, as a means to working around the diffraction limit, depends on both specific microscopy protocols and extensive software post-exposure.
Dr. Shao and his team, led by Mats Gustafsson, have made a number of discoveries using SI. “We have shown that, when using SI in a linear regime, that the resolution limit of the wide-field microscopy can be improved by a factor of two both laterally and axially.
“Data acquisition, however, is not quick enough to achieve live imaging. The reason for this is that reconstructing a 2-D image requires multiple exposures under different illumination patterns.” He added that 15 images are required per every 2-D section in 3-D imaging. Mechanical pattern switching is also slow.
Dr. Shao also spoke about an SI microscope developed to use a ferroelectric liquid crystal-based spatial light modulator as the pattern generator. “The image-acquisition speed is greatly enhanced by the greater than 1 KHz pattern-switching speed of the spatial light modulator and the total internal reflection mode that allows 2-D imaging and thus only nine exposures per time point. As a result, the microscope is capable of 100 nm resolution at frame rates up to 11 Hz for several hundred time points.”
Dr. Shao demonstrated the speed and resolution of the microscope by imaging tubulin and kinesin dynamics in live Drosophila melanogaster S2 cells.
“Speed is an issue, since to keep up with fast moving molecules inside cells requires fast cameras with high resolution. The dynamic nature is also a challenge—how do you maintain the level of fluorescence without bleaching the phosphores. The additional problem of high resolution applies to both issues. And another obstacle is how one can keep the cells happy while being imaged, which involves mounting schemes for temperature control (mammalian cells are happy around 37ºC; S2 cells are happy at RT) and being careful about phototoxicity, i.e., not too high-intensity excitation light should be allowed.”
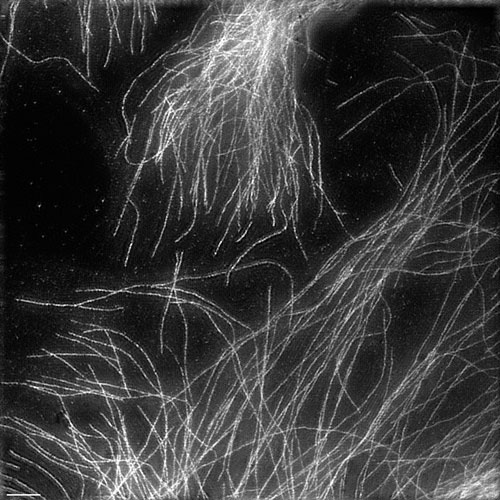
Fixed HeLa cells in which tubulin antibody is stained [Howard Hughes Medical Institute]
Fluorescent Cell-Based Analyses
Dr. Persmark gave an overview of several of Life Technologies’ recently launched reagents for fluorescent cell-based analyses at the “Immunology” meeting. “In cellular imaging, we are focusing efforts on probes and technologies to monitor cell structure and function—some entirely novel and some that draw heavily on our 30-year history, such as Alexa Fluor® dyes. However, the Alexa Fluor dyes are also being used in completely new technology applications, such as the Click-based assays.”
In order to deal with some of the challenges inherent in imaging live cells, Life Technologies has developed BacMam technology, which is based on an insect virus (baculovirus) to efficiently deliver and express genes in mammalian cells.
Premo™ FUCCI Cell Cycle Sensor combines Cdt1 and geminin FP constructs with the BacMam gene delivery system. The Premo sensor is based on the two-color FUCCI sensor developed by Atsushi Miyawaki and colleagues at Riken; depending on the mitotic phase the sensor fluoresces in red or green (or yellow at the G1/S interphase), explained Dr. Persmark.
“Packaging in BacMam particles enables researchers to study the mitotic state of individual cells, including stem cells and neuronal cells that traditionally are challenging to label.”
The BacMam platform 2.0 includes elements that enhance transduction efficiency and expression levels: a pseudotyped capsid protein for more efficient cell entry and genetic elements (enhanced CMV promoter and Woodchuck Post-transcriptional Regulatory Element), that boost expression levels. Baculoviruses do not replicate in mammalian cells and thus have an excellent safety profile and lack cytopathic effects on cells, Dr. Persmark added.
“BacMam reagents have been used in cell-based assays, live-cell imaging, stem cell biology, and many other applications. Owing to the high transduction efficiency and lack of cytotoxicity, it is easy to perform experiments in more biologically relevant cellular models—for example, stem cells, neurons, and primary cells.” Dr. Persmark added that for multicomponent systems that require stoichiometric expression, the transient expression and ability to control expression by simply varying the dose enables analysis and even screening of targets that have proven intractable using stable cell lines.
Dr. Christensen noted that many associate live-cell imaging with high-content analysis in terms of microscopy. “Working down to the sub-cellular level, you can get valuable insight to direct cellular response and intracellular location of functional activity,” she said.
“On the other hand, microscopy lacks a great deal of biological relevance. How a cell responds on a slide can be vastly different from how a cell responds in a living organism. Characterization of cellular responses can easily be accomplished at a whole-animal level through noninvasive optical-imaging techniques.”
Optical-imaging technologies rely on light producing optical reporters such as luciferase and fluorescent proteins, fluorescent dyes, and conjugates. “Genes encoding luciferase and fluorescent proteins can be engineered into cells—for example, cancer cell lines and infectious disease agents or animals such as transgenic mice and rats—to enable them to produce light that can then be visualized through the tissues of a live animal using specialized imaging equipment and software designed and built by the company,” said Dr. Christensen.
“These highly sensitive dual bioluminescence and fluorescence imaging systems allow significantly fewer animals to be used due to the generation of superior data and better biostatistics. Optical imaging enables one to characterize functional processes and responses to therapeutic treatment longitudinally and translationally.”
Dr. Christensen noted that there isn’t any one technology that’s optimal for live-cell imaging. “The combination of microscopic and macroscopic techniques are required to fully understand the complete biology at hand,” she said. Live-cell imaging, however, brings more relevance to biological processes in real time. Identification of an event at a specific time can be easily done on fixed cells, but characterization of an event more often needs real-time monitoring of the biological process.”


